dnorm(x, mean = 0, sd = 1, log = FALSE)
pnorm(q, mean = 0, sd = 1, lower.tail = TRUE, log.p = FALSE)
qnorm(p, mean = 0, sd = 1, lower.tail = TRUE, log.p = FALSE)
rnorm(n, mean = 0, sd = 1)x3-Functions Everywhere
Applied Statistics – A Practical Course
2026-01-28
Functions bring life to the R language
sin(x), log(x), plot(x, y), summary(x), anova(lm.object), mean(x), monod(S, vmax, ks), simulate_phytoplankton(N, P, T, Zoo, ...)
Functions in R
- have a name, followed by parenthesis ()
- can have 1, 2 or more arguments (or no argument)
- usually return something (an object)
- can have side-effects (e.g. plotting)
What are functions
Parentheses and arguments
- all functions are followed by parentheses and arguments
- functions:
log(x)par() par <- c(a=5, b=3)
\(\rightarrow\) here, par is a variable, c() a function
Return value and/or side effect
sin(x),log(x),mean(x)are functions with return valueprint(x),plot(x, y)are functions with side effecthist(x)is a function with both, side effect and return value
Predefined and user-defined functions
- predefined: available in R
- user defined: users become programmers
Arguments of functions
Usage
| x, q | vector of quantiles. |
| p | vector of probabilities. |
| n | number of observations. If length(n) > 1, the length is taken to be … |
| log.p | if TRUE, probabilities p are given as log(p). |
| lower.tail | if TRUE (default), … |
Arguments
- required arguments: have no default
- optional arguments: have default values
- named arguments: argument mathing with
=allows to specify arguments in arbitrary order - argument order: arguments can occur without names when in defined order
- “…”: dots-arguments are passed down to other called functions
Examples
The ellipsis argument
- Some functions have a … argument, called “ellipsis”.
- This means that additional arguments are passed to other functions.
- Makes R flexible and extensible, but is sometimes tricky.
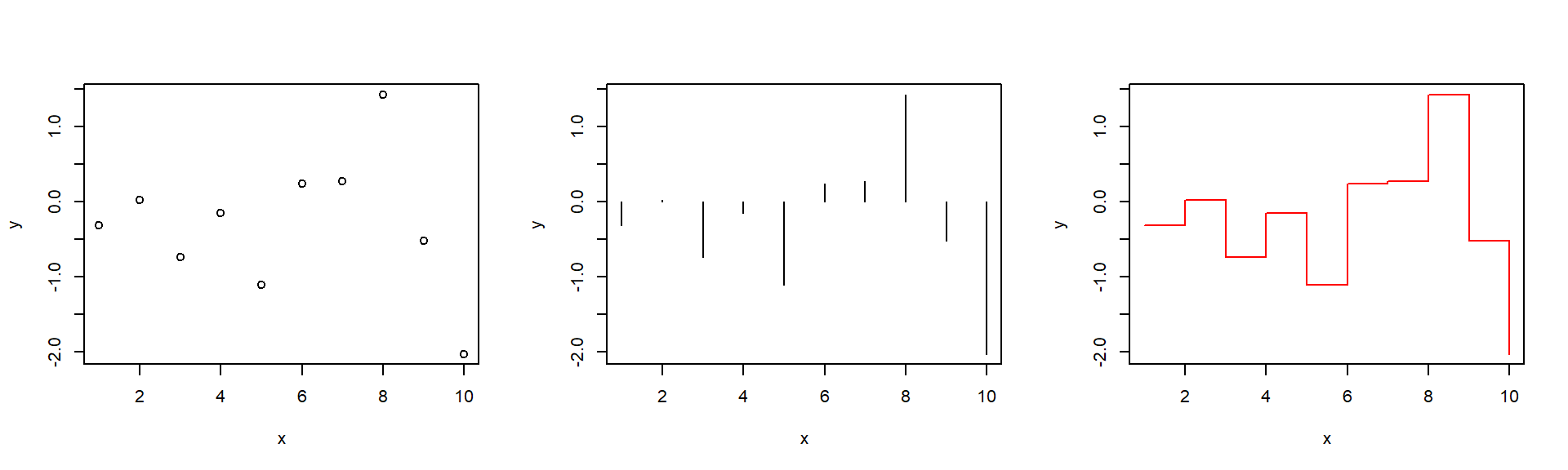
plot.default
Object orientation
plotis a generic function- works automagic differently for different classes of objects
plot.defaultis the basic function...see?parfor additional graphical parameters, e.g.:
col |
color |
bg |
background color for two-color symbols |
pch |
symbol (plotting character) |
cex |
size of symbol (character extension) |
lty |
line type |
lwd |
line width |
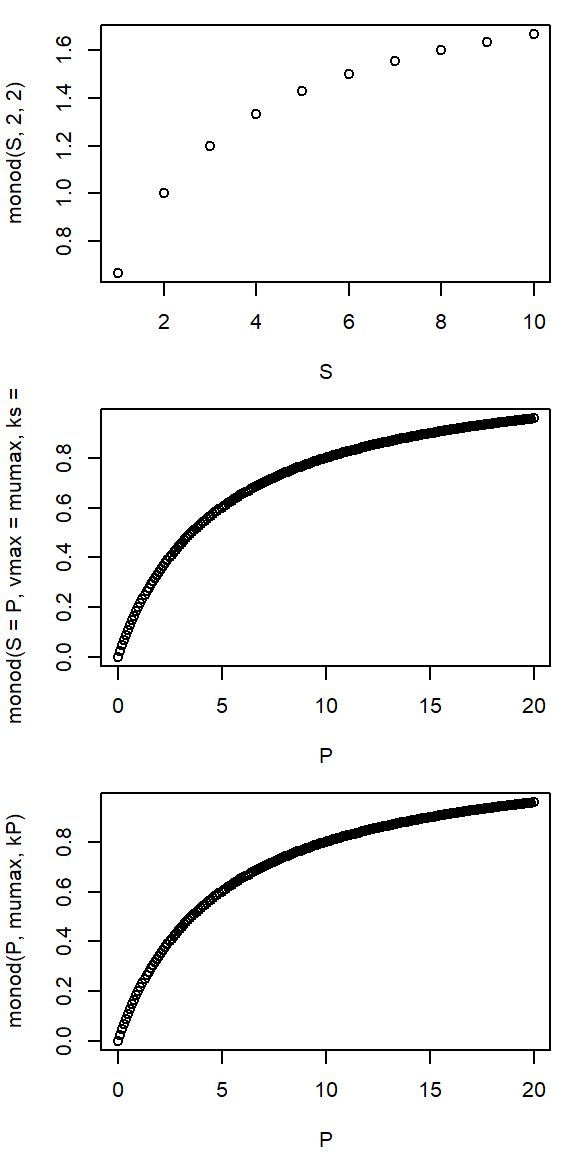
A user-defined Monod function
- describes substrate dependence of biochemical turnover
- widely used in biochemistry and in models
- e.g. organic matter turnover in wastewater treatment
\[ v = \frac{v_{max} \cdot S}{k_S + S} \]
par(mar=c(4,4,1,1))
par(mfrow=c(3, 1))
monod <- function(S, vmax, ks) {
vmax * S / (ks + S)
}
S <- 1:10
P <- seq(0, 20, 0.1)
kP <- 5; mumax <- 1.2;
## different ways to call the function
plot(S, monod(S, 2, 2)) # simple call
plot(P, monod(S=P, vmax=mumax, ks=kP)) # named arguments
plot(P, monod(P, mumax, kP)) # argument position- names of caller and function can be different
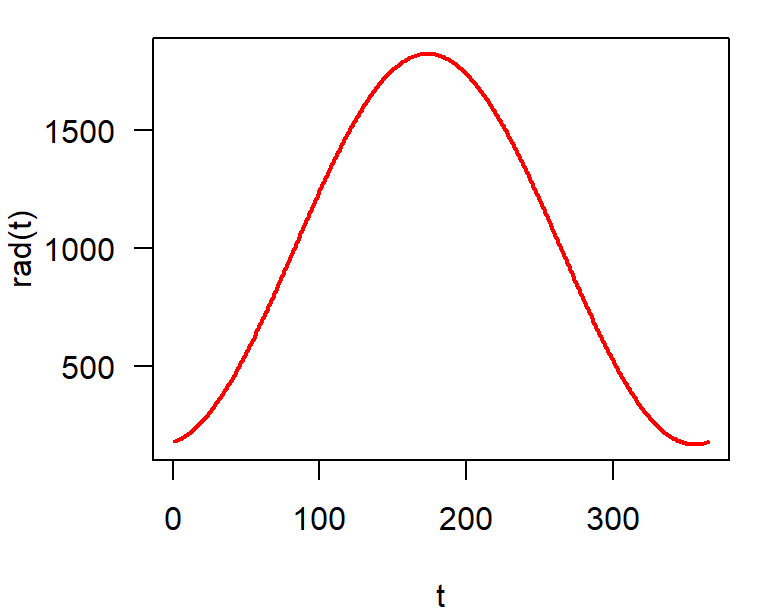
Seasonal Light Intensity in Dresden
\[ I_t = 997 - 816 \cos(2 \pi t / 365) + 126 \sin(2 \pi t / 365) \]
Functions as a knowledge base
- put knowledge in function and use it
- forget what is inside
Oxygen saturation in fresh and sea water
\[ c_{O_2, 100\%} = ... ? \]
o2sat <- function(t) {
K <- t + 273.15 # Celsius to Kelvin
exp(-139.34411 + (157570.1/K) - (66423080/K^2) +
(1.2438e+10/K^3) - (862194900000/K^4))
}
o2sat(20)[1] 9.092426
A more precise formula is found in package marelac
consult ?gas_O2sat for citations.
Local and global variables
Variables in a function are local:
- not visible from outside.
- no collisions with existing variables in the calling environment
Lexical Scoping
- functions can see variables of the calling function
- useful for interactive work
- dangerous for (exported) functions in packages
- except in special cases, e.g. for functions within functions
Local and global variables II
Now outcomment:
and try again.
Exercise
Logistic growth
The logistic growth function describes saturated growth of a population abundance \(N_t\), dependent of an initial value \(N_0\), growth rate \(r\) and carrying capacity \(K\).
\[ N_t = \frac{K N_0 e^{rt}}{K + N_0 (e^{rt}-1)} \]
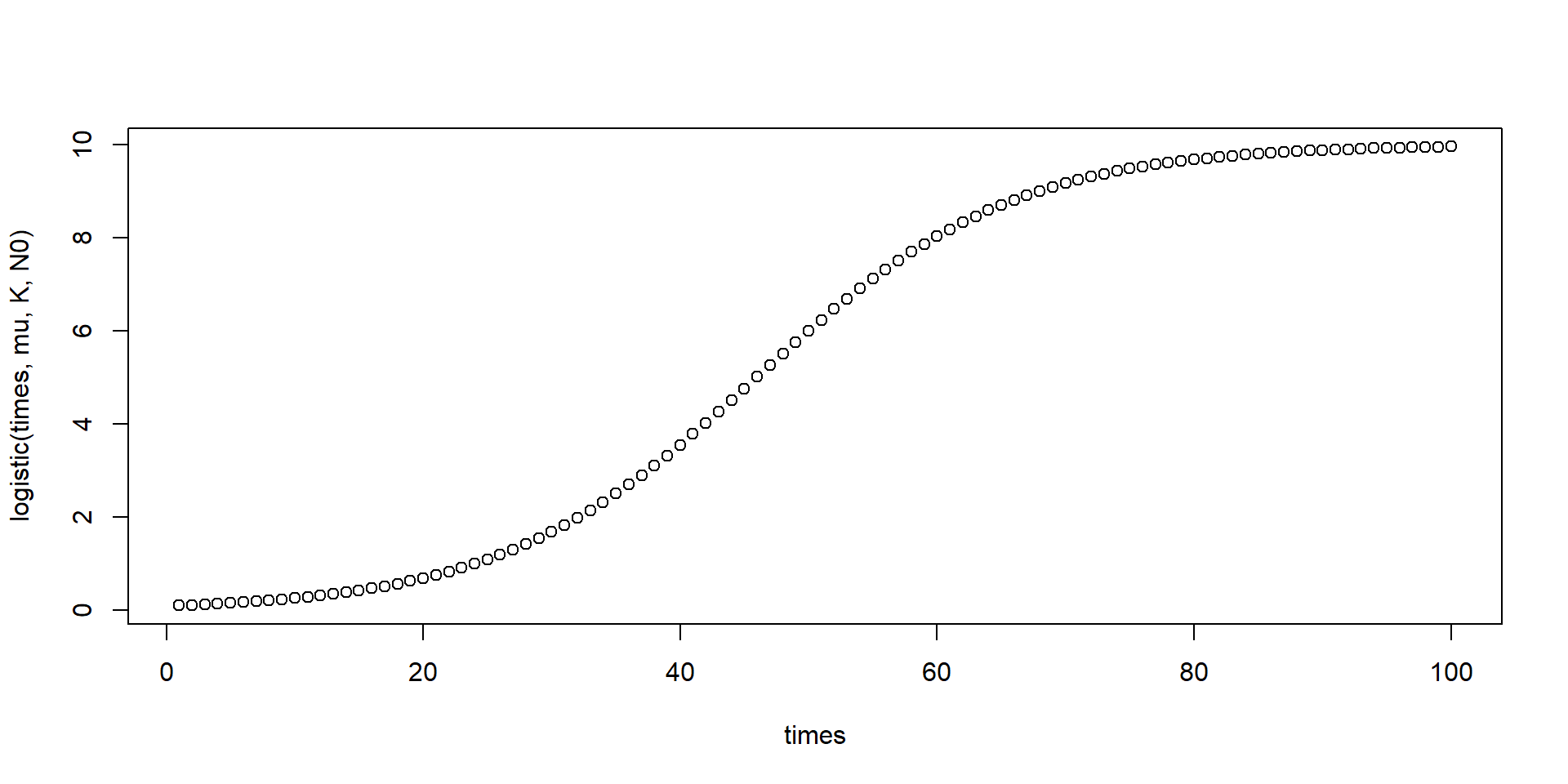
Functional response types in Ecology
- Holling type I \(P = \min(k \cdot N, P_{max})\)
- Holling type II \(P = \frac{\alpha N}{1 + \alpha H N}\)
- Holling type III \(P = \frac{\alpha N^b}{1 + \alpha H N^b}\)
with
| \(P\) | predation rate |
| \(N\) | abundance of prey |
| \(P_{max}\) | maximum predation rate |
| \(k\) | a constant |
| \(\alpha\) | attack rate |
| \(H\) | handling time |
| \(b\) | exponent \(>1\) |
- Write a function for each functional reponse type and plot it.
- Write a universal function for all types.
Further Reading
More presentations
Manuals
More details in the official R manuals, especially in An Introduction to R
Videos
Many videos can be found on Youtube, at the Posit webpage and somewhere else.