A data frame is split into a list of data subsets defined by multiple groups.
Usage
multisplit(data, grouping, drop = TRUE, sep = ":", ...)
# S4 method for class 'data.frame,formula'
multisplit(data, grouping, drop = TRUE, sep = ":", ...)
# S4 method for class 'data.frame,character'
multisplit(data, grouping, drop = TRUE, sep = ":", ...)
# S4 method for class 'data.frame,factor'
multisplit(data, grouping, drop = TRUE, sep = ":", ...)
# S4 method for class 'data.frame,list'
multisplit(data, grouping, drop = TRUE, sep = ":", ...)
# S4 method for class 'ANY,ANY'
multisplit(data, grouping, drop = TRUE, sep = ":", ...)Arguments
- data
data frame, matrix or vector containing several subsets of data
- grouping
either a character vector containing the names of the grouping variables or a model formula specifying dependent, independent and grouping variables in the form:
dependent ~ independent | group1 + group2 + .... It may also be a factor or list of factors as insplit.- drop
if drop is TRUE, unused factor levels are dropped from the result. The default is to drop all factor levels.
- sep
string to construct the new level labels by joining the constituent ones.
- ...
other parameters passed to
split, see details.
Value
list containing data frames of the data subsets as its elements. The components of the list are named by their grouping levels.
Details
This function is wrapper around split with
different defaults, slightly different behavior, and methods for additional
argument classes. multisplit returns always a data frame.
Examples
data(bactgrowth)
## simple method
spl <- multisplit(bactgrowth, c("strain", "conc", "replicate"))
## preferred method
spl <- multisplit(bactgrowth, value ~ time | strain + conc + replicate)
## show what is in one data set
spl[[1]]
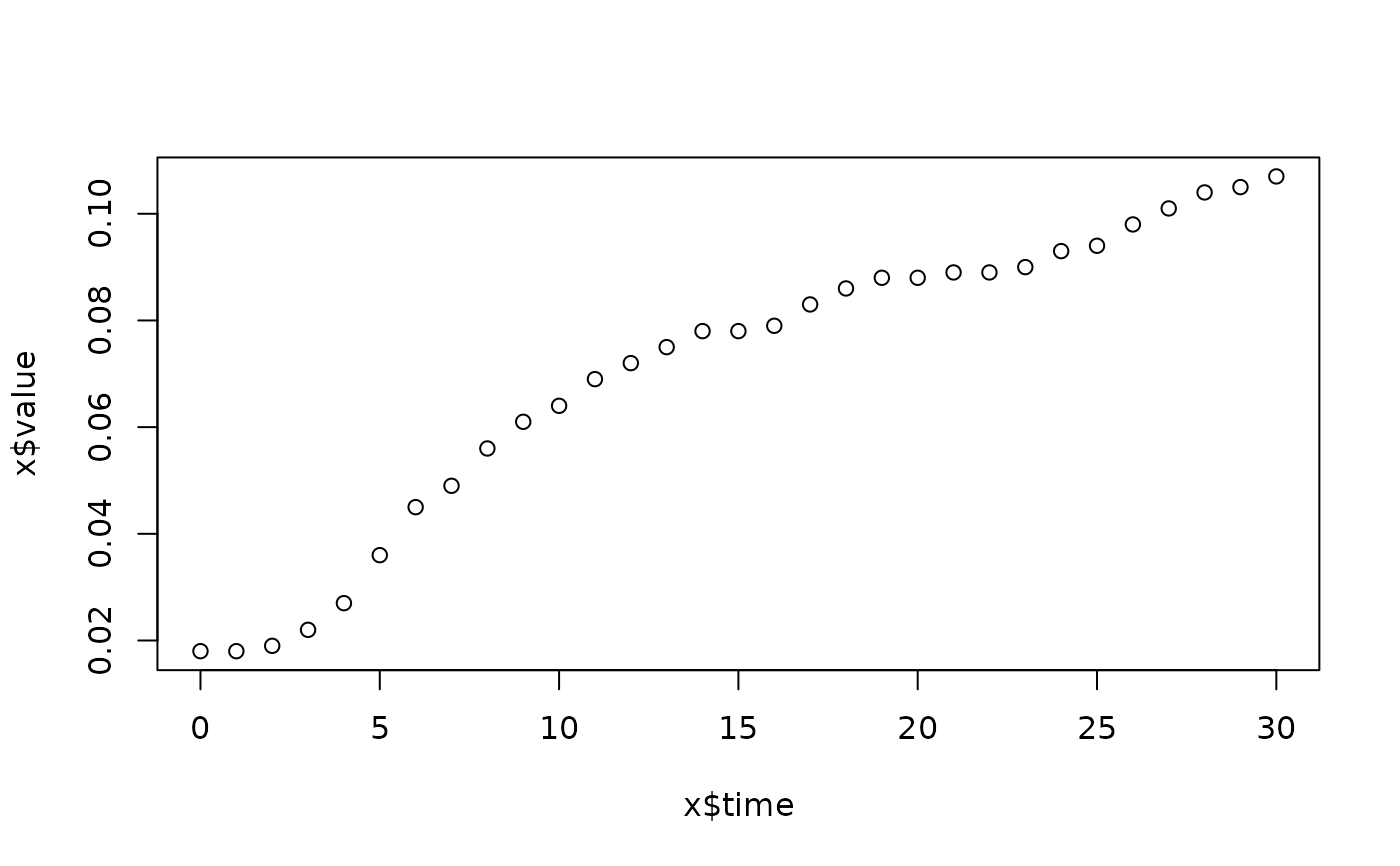
#> time value
#> 1117 0 0.018
#> 1118 1 0.018
#> 1119 2 0.019
#> 1120 3 0.022
#> 1121 4 0.027
#> 1122 5 0.036
#> 1123 6 0.045
#> 1124 7 0.049
#> 1125 8 0.056
#> 1126 9 0.061
#> 1127 10 0.064
#> 1128 11 0.069
#> 1129 12 0.072
#> 1130 13 0.075
#> 1131 14 0.078
#> 1132 15 0.078
#> 1133 16 0.079
#> 1134 17 0.083
#> 1135 18 0.086
#> 1136 19 0.088
#> 1137 20 0.088
#> 1138 21 0.089
#> 1139 22 0.089
#> 1140 23 0.090
#> 1141 24 0.093
#> 1142 25 0.094
#> 1143 26 0.098
#> 1144 27 0.101
#> 1145 28 0.104
#> 1146 29 0.105
#> 1147 30 0.107
summary(spl[[1]])
#> time value
#> Min. : 0.0 Min. :0.01800
#> 1st Qu.: 7.5 1st Qu.:0.05250
#> Median :15.0 Median :0.07800
#> Mean :15.0 Mean :0.07035
#> 3rd Qu.:22.5 3rd Qu.:0.08950
#> Max. :30.0 Max. :0.10700
## use factor combination
spl[["D:0:1"]]
#> time value
#> 1117 0 0.018
#> 1118 1 0.018
#> 1119 2 0.019
#> 1120 3 0.022
#> 1121 4 0.027
#> 1122 5 0.036
#> 1123 6 0.045
#> 1124 7 0.049
#> 1125 8 0.056
#> 1126 9 0.061
#> 1127 10 0.064
#> 1128 11 0.069
#> 1129 12 0.072
#> 1130 13 0.075
#> 1131 14 0.078
#> 1132 15 0.078
#> 1133 16 0.079
#> 1134 17 0.083
#> 1135 18 0.086
#> 1136 19 0.088
#> 1137 20 0.088
#> 1138 21 0.089
#> 1139 22 0.089
#> 1140 23 0.090
#> 1141 24 0.093
#> 1142 25 0.094
#> 1143 26 0.098
#> 1144 27 0.101
#> 1145 28 0.104
#> 1146 29 0.105
#> 1147 30 0.107
summary(spl[["D:0:1"]])
#> time value
#> Min. : 0.0 Min. :0.01800
#> 1st Qu.: 7.5 1st Qu.:0.05250
#> Median :15.0 Median :0.07800
#> Mean :15.0 Mean :0.07035
#> 3rd Qu.:22.5 3rd Qu.:0.08950
#> Max. :30.0 Max. :0.10700
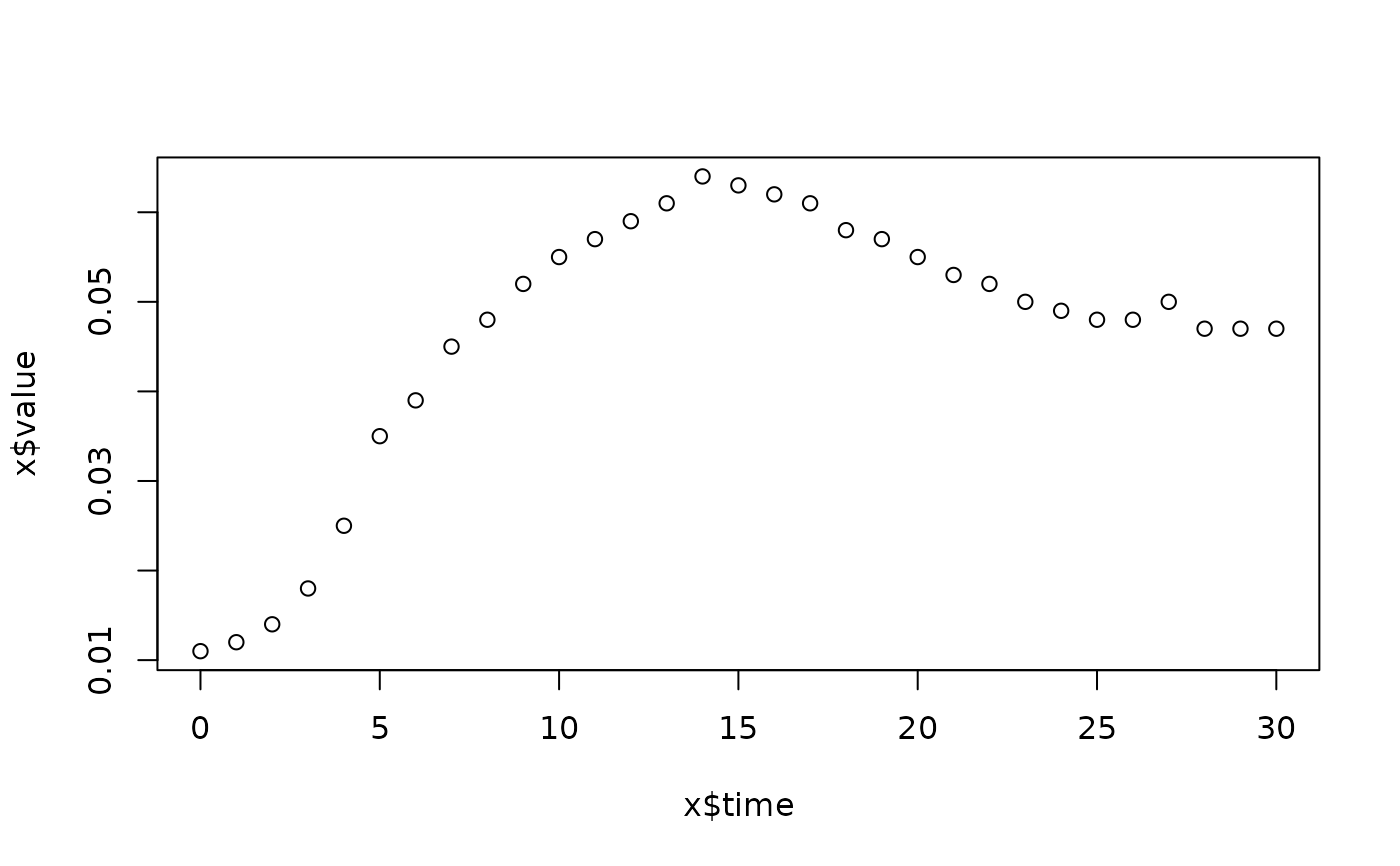
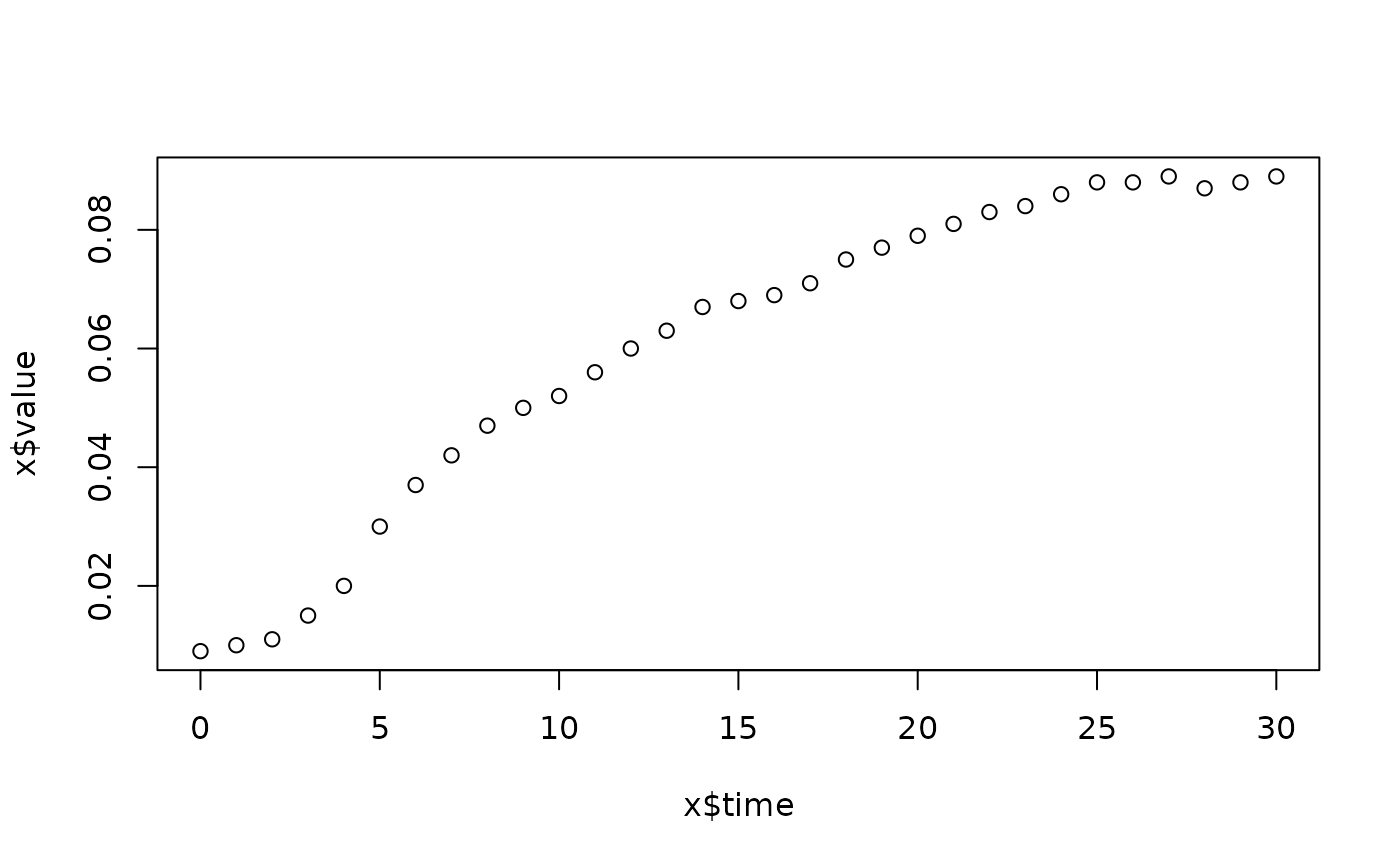
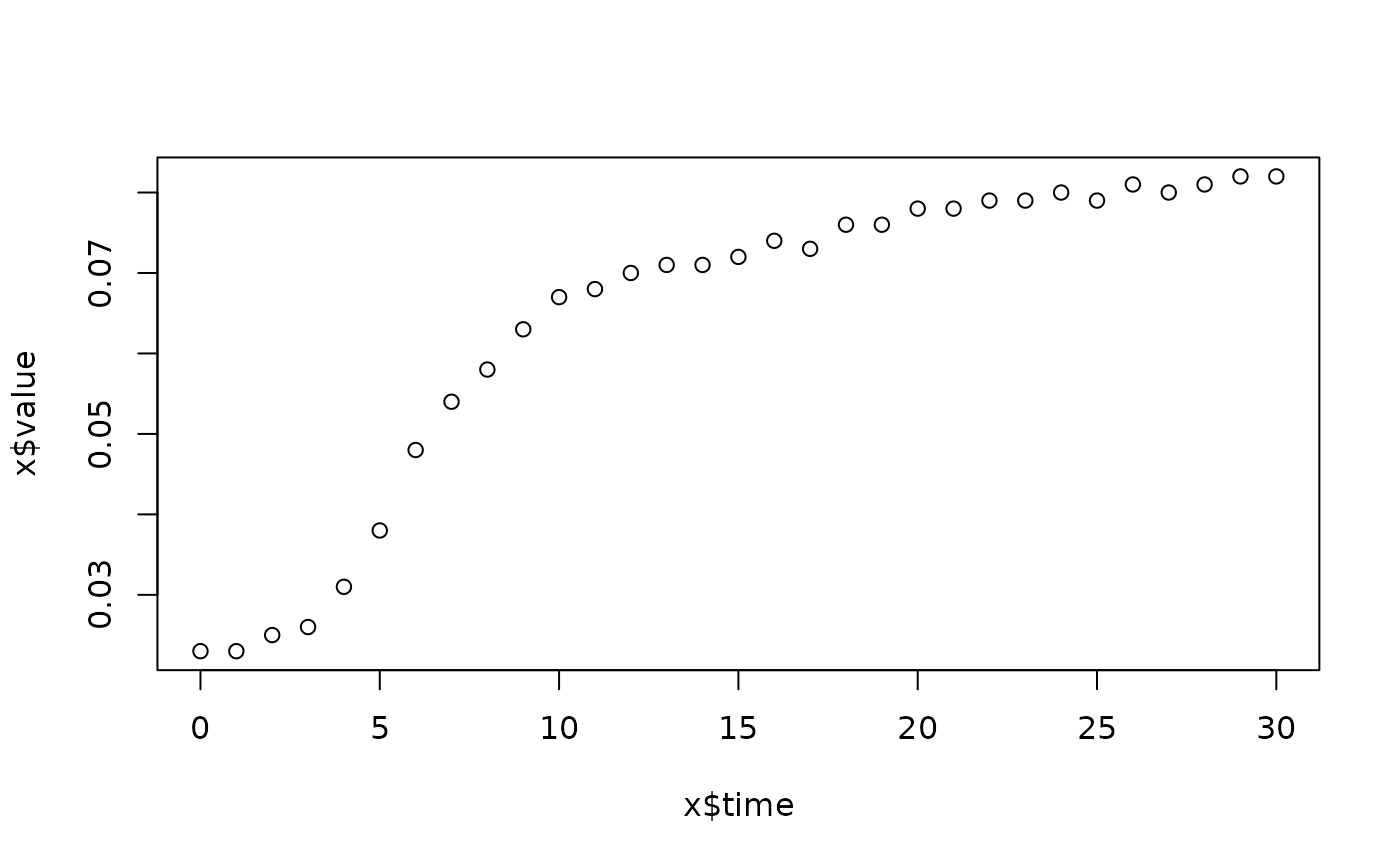
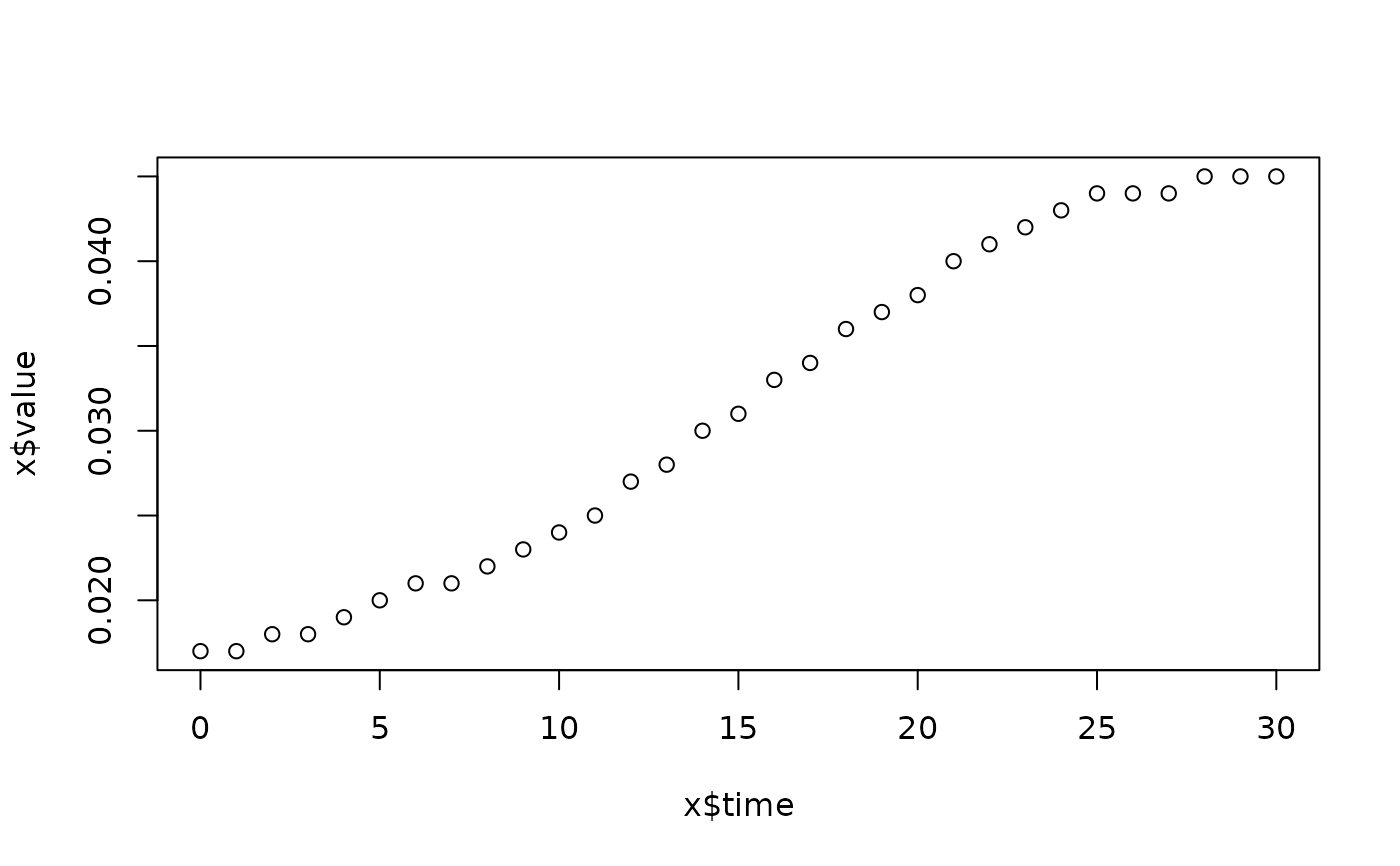
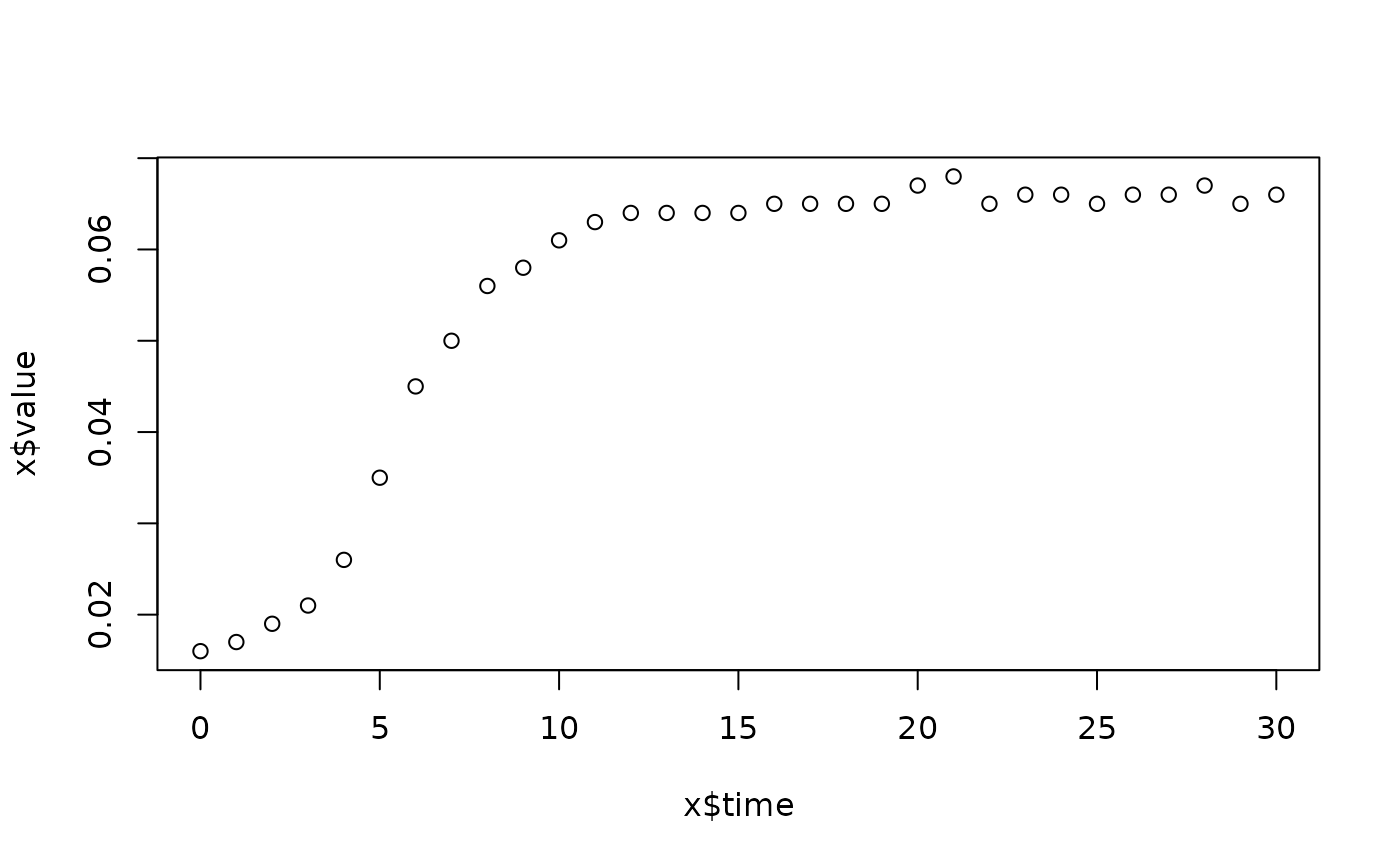
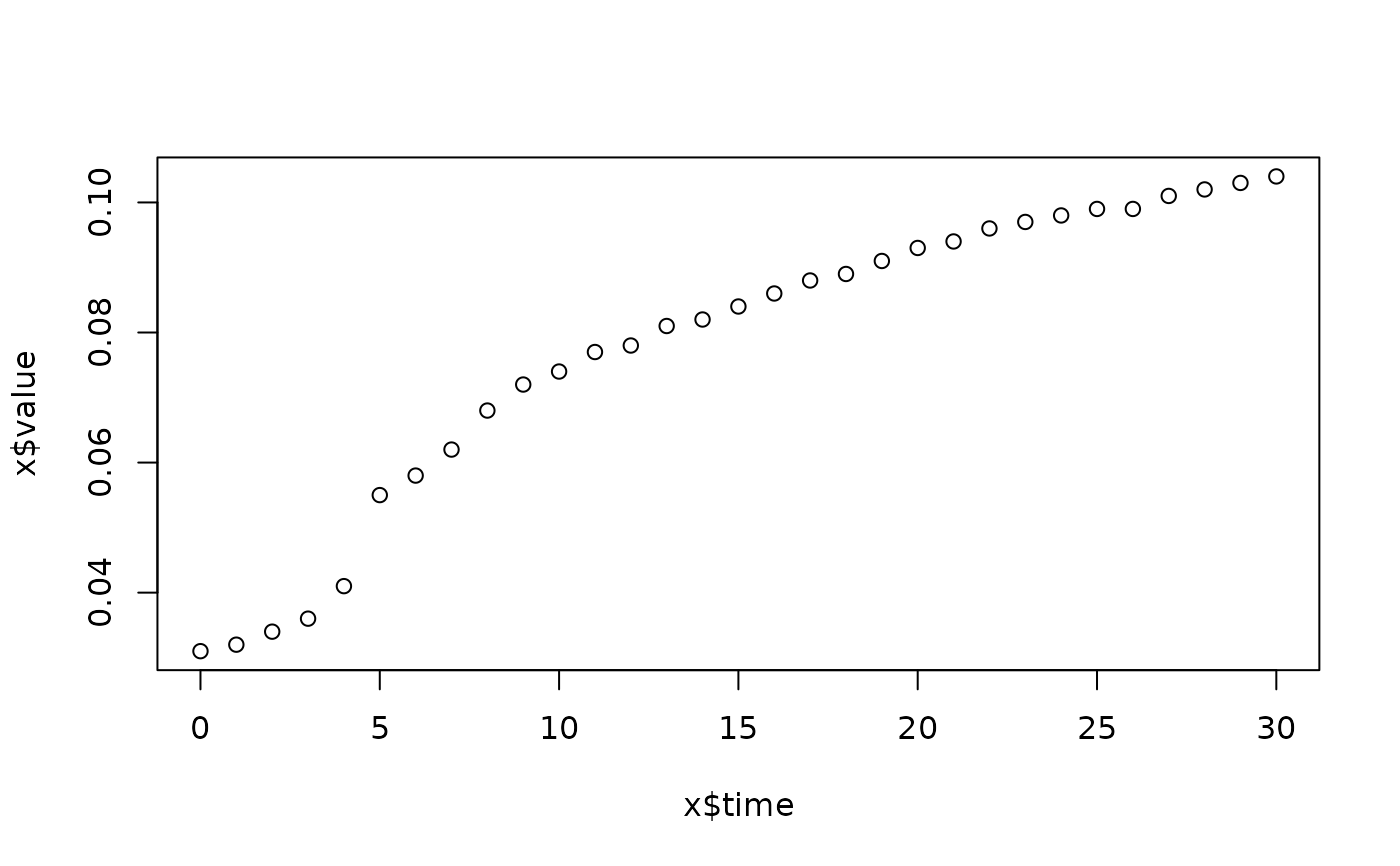
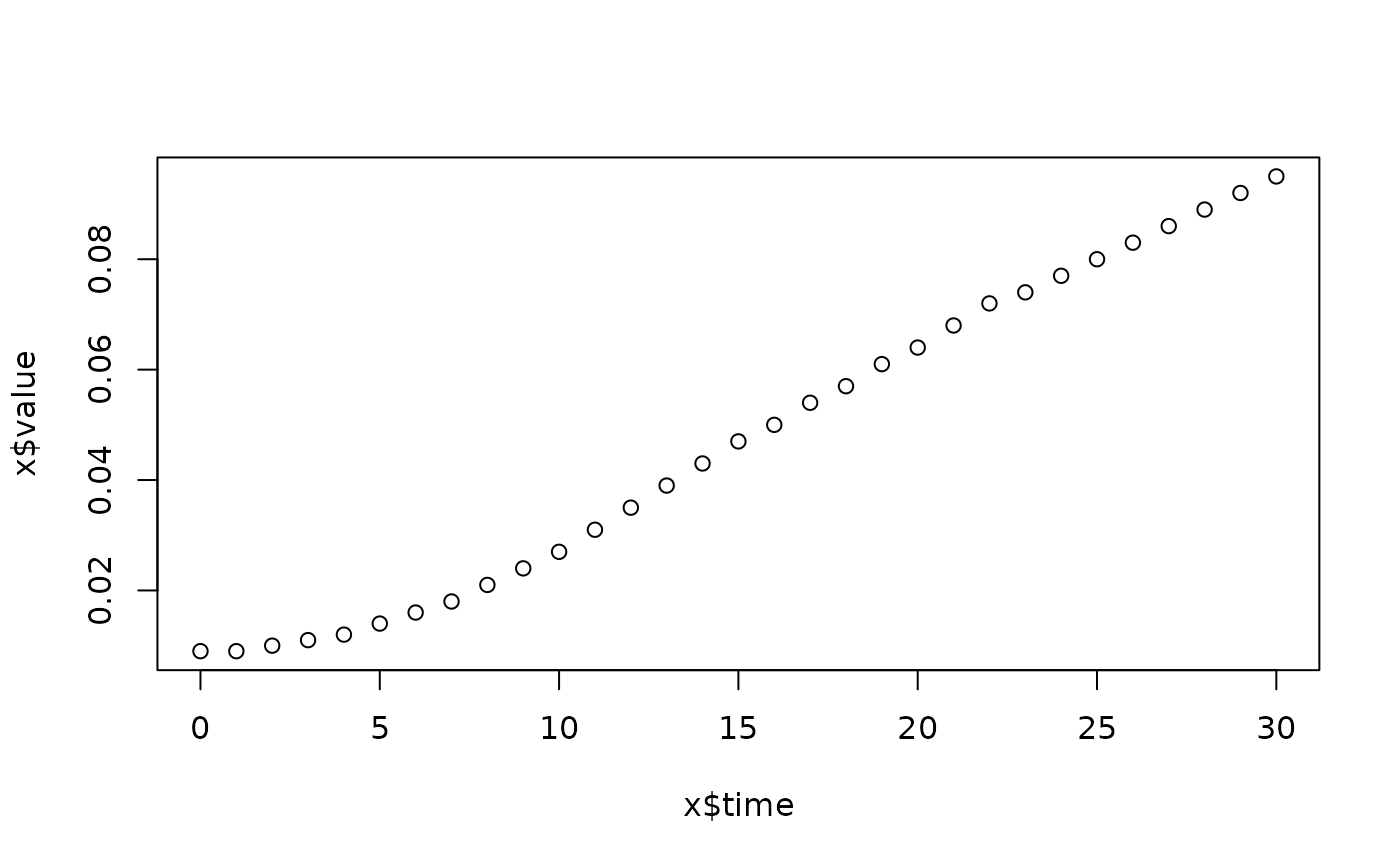
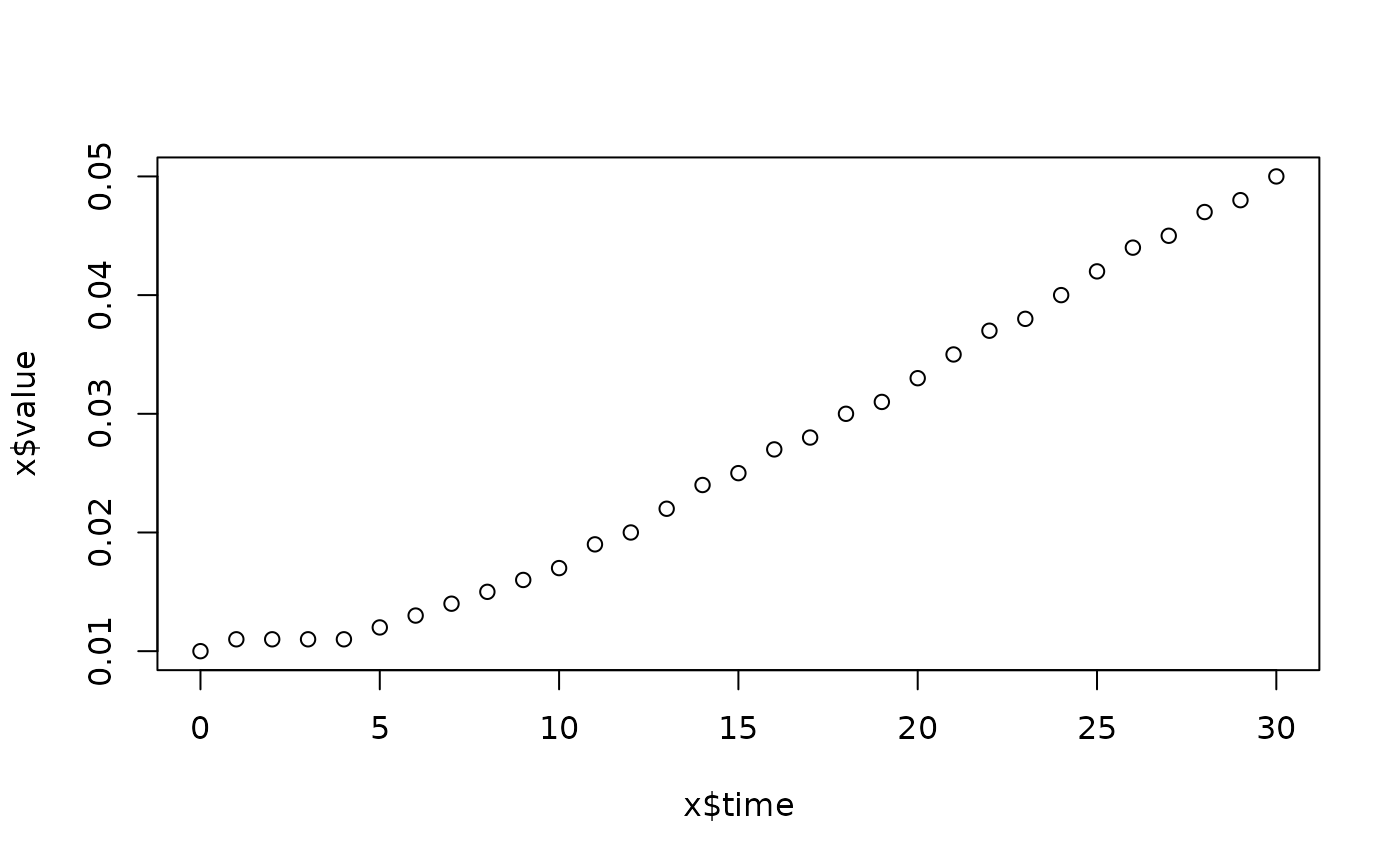
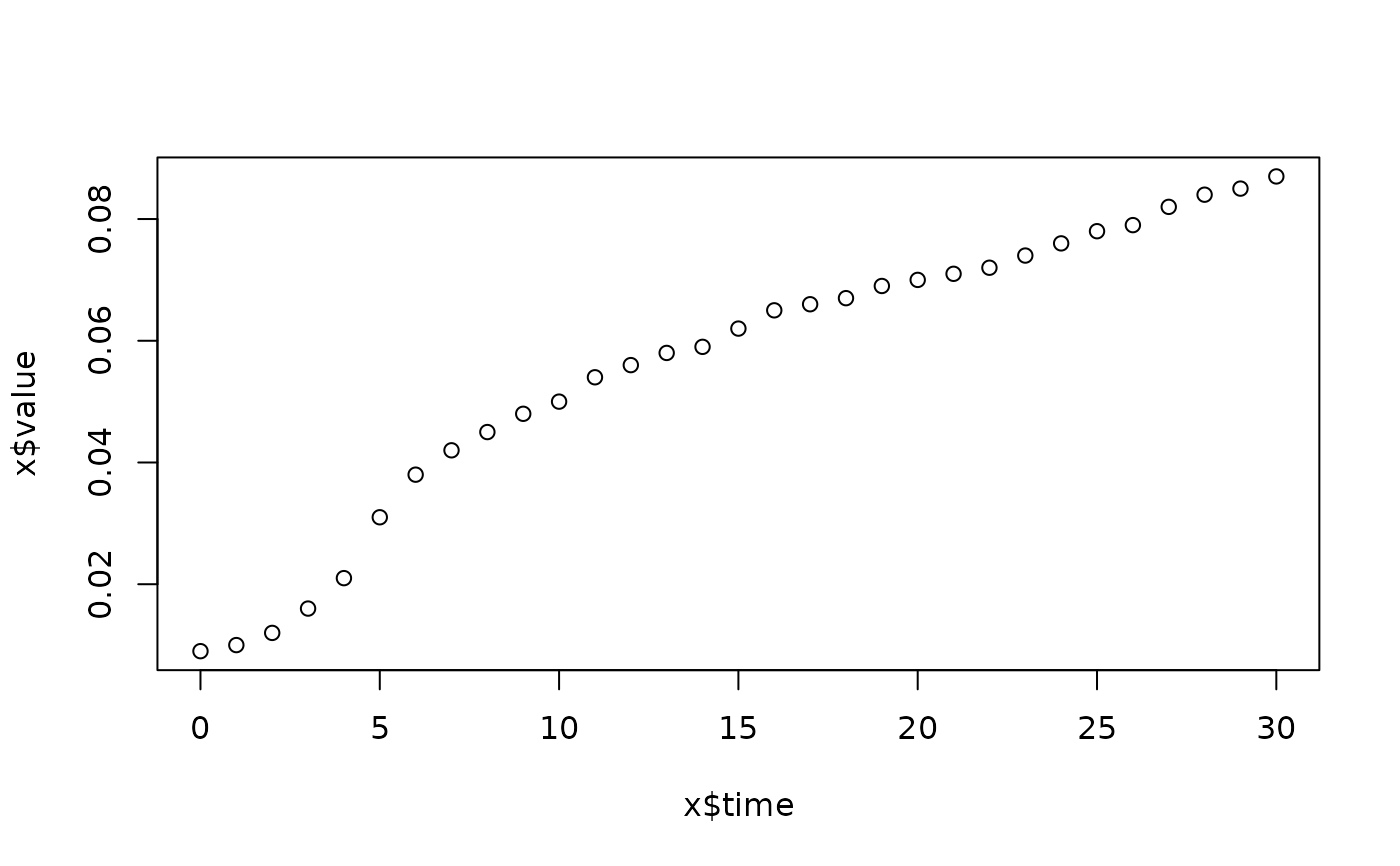
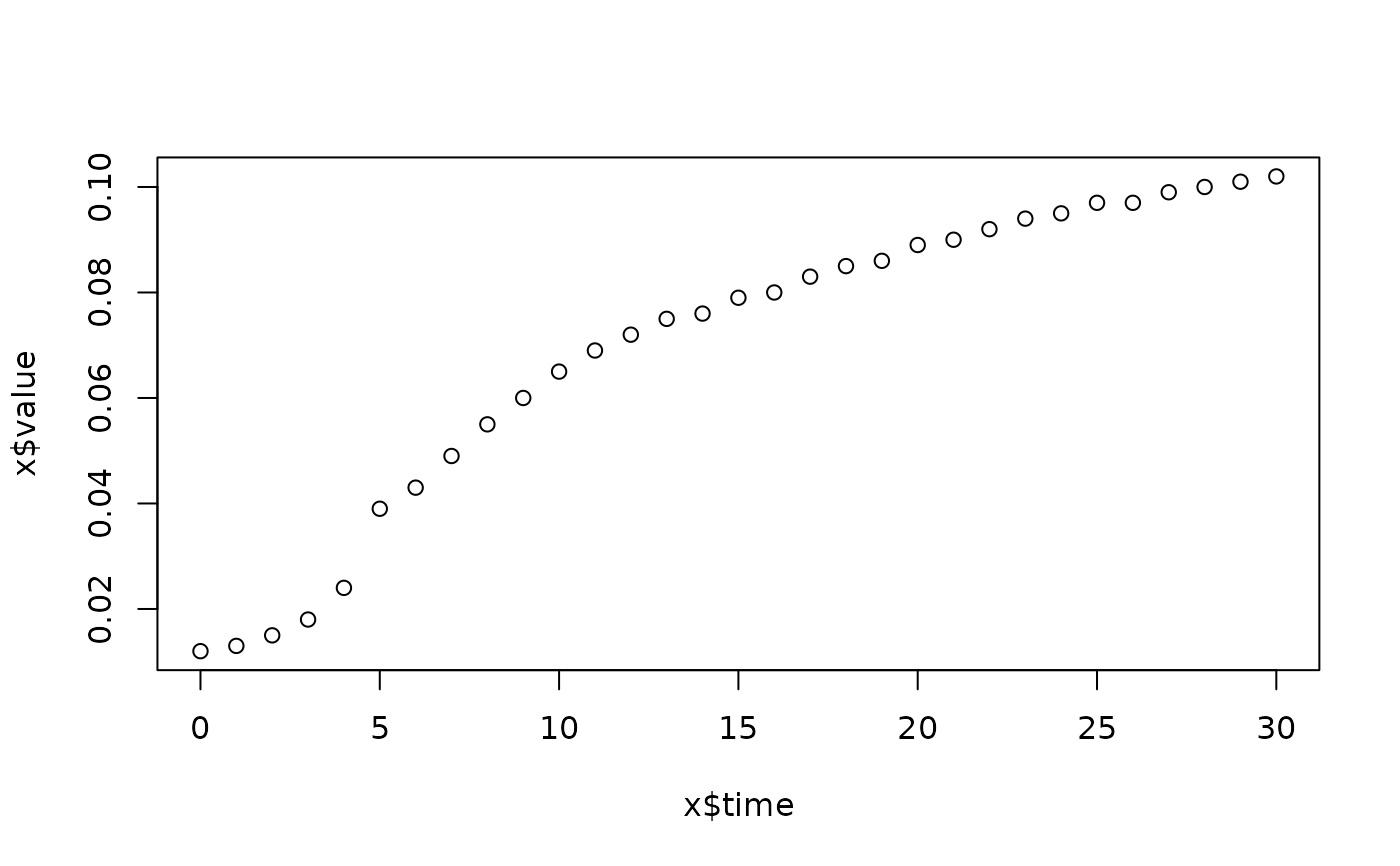
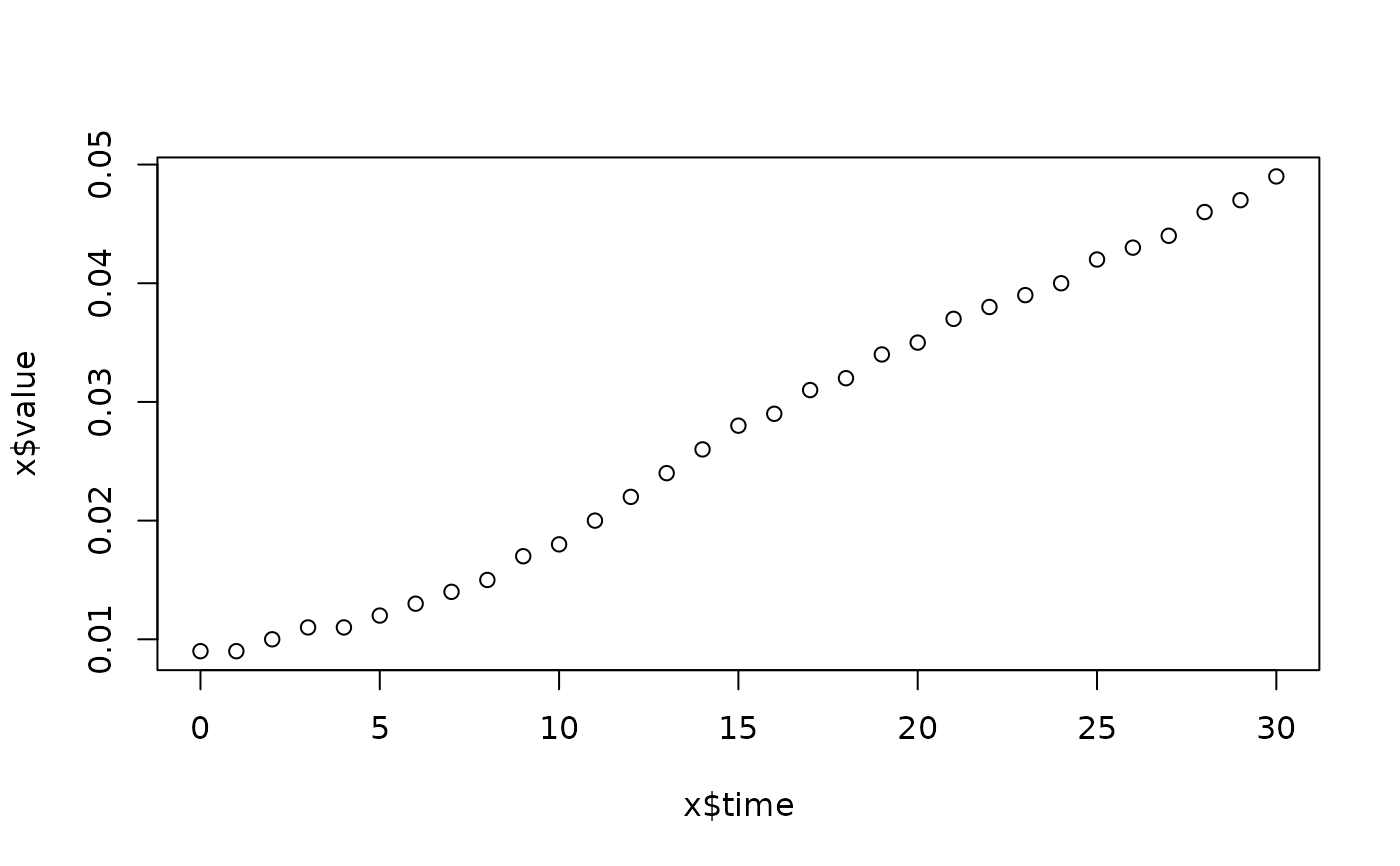
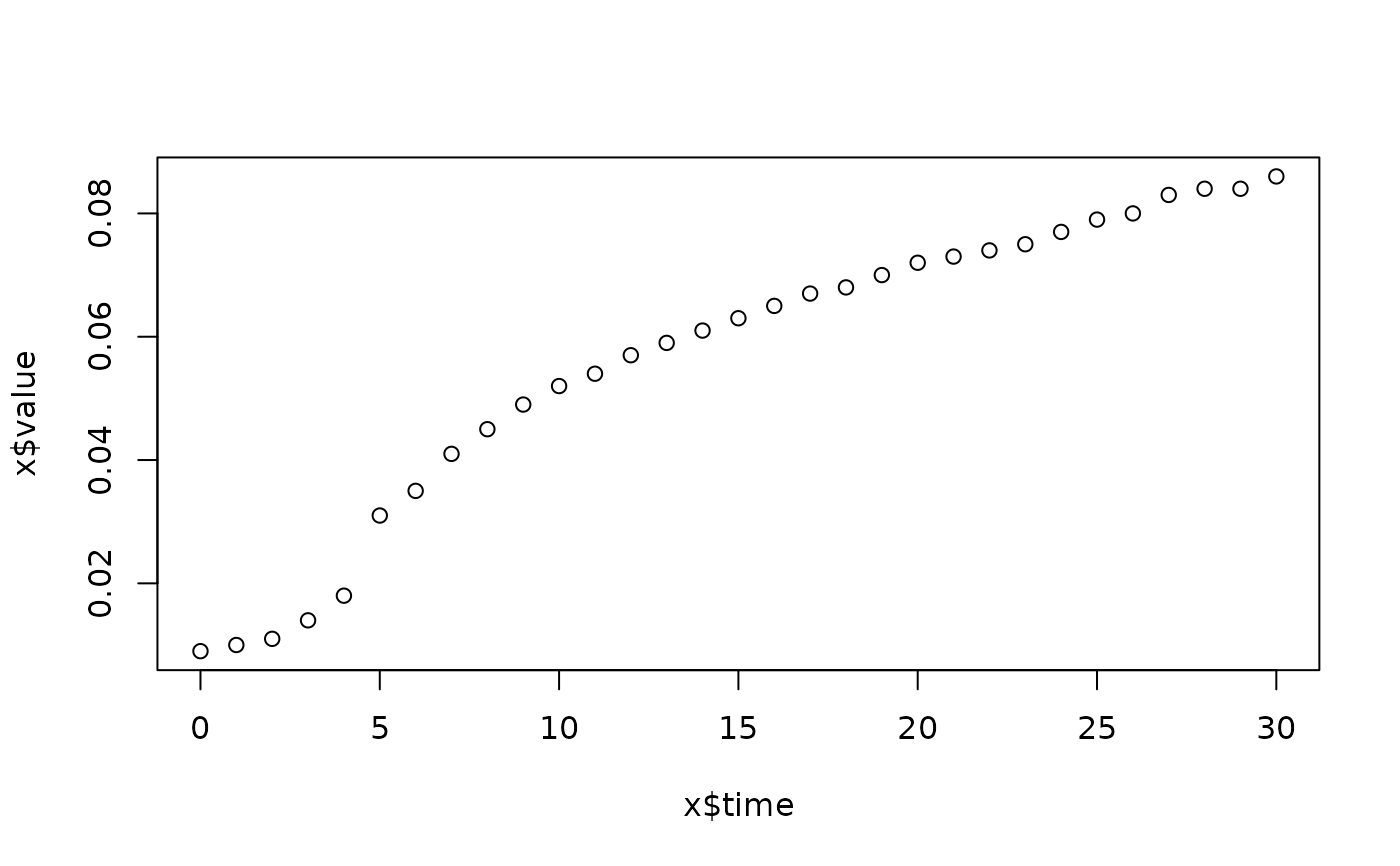
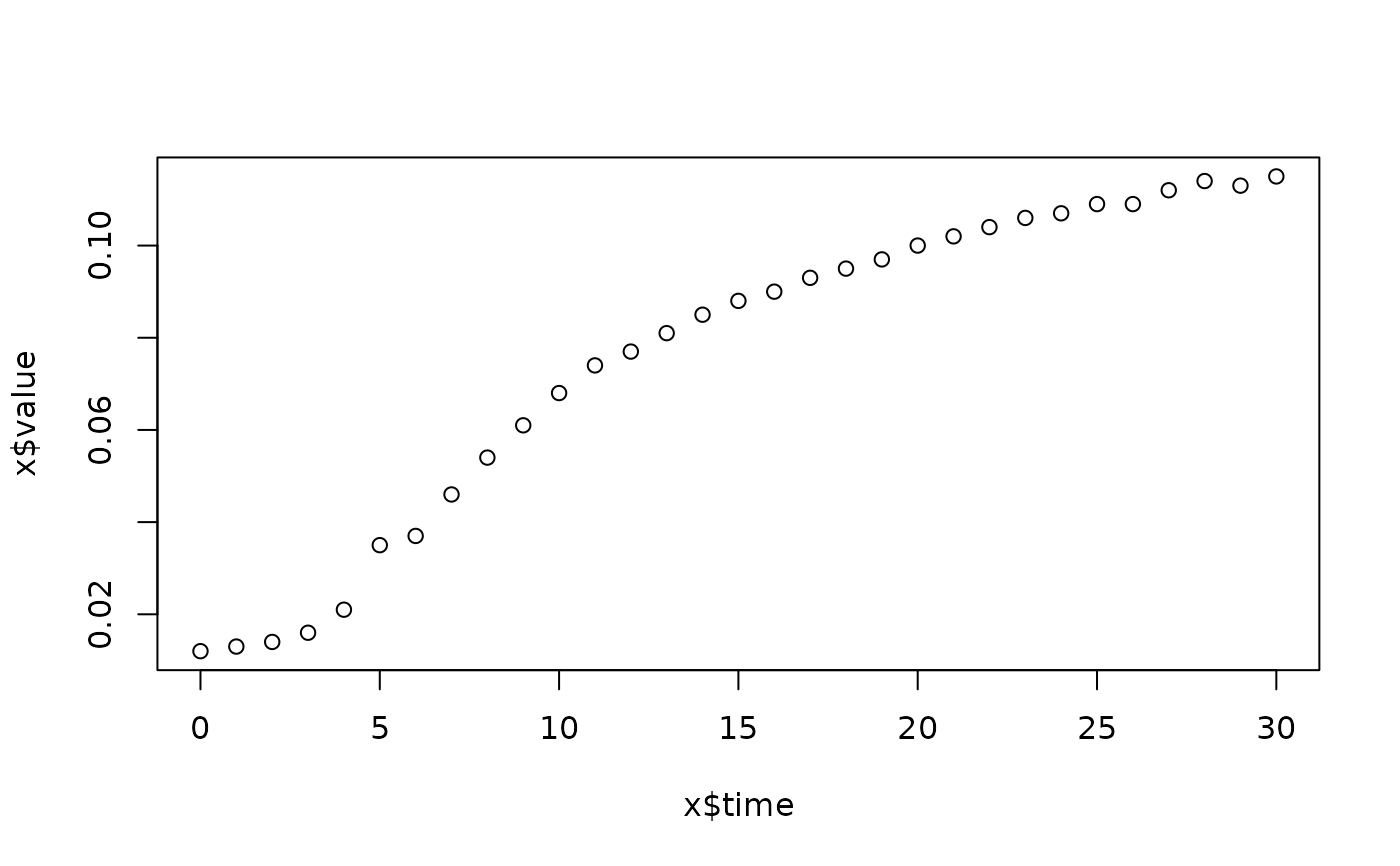
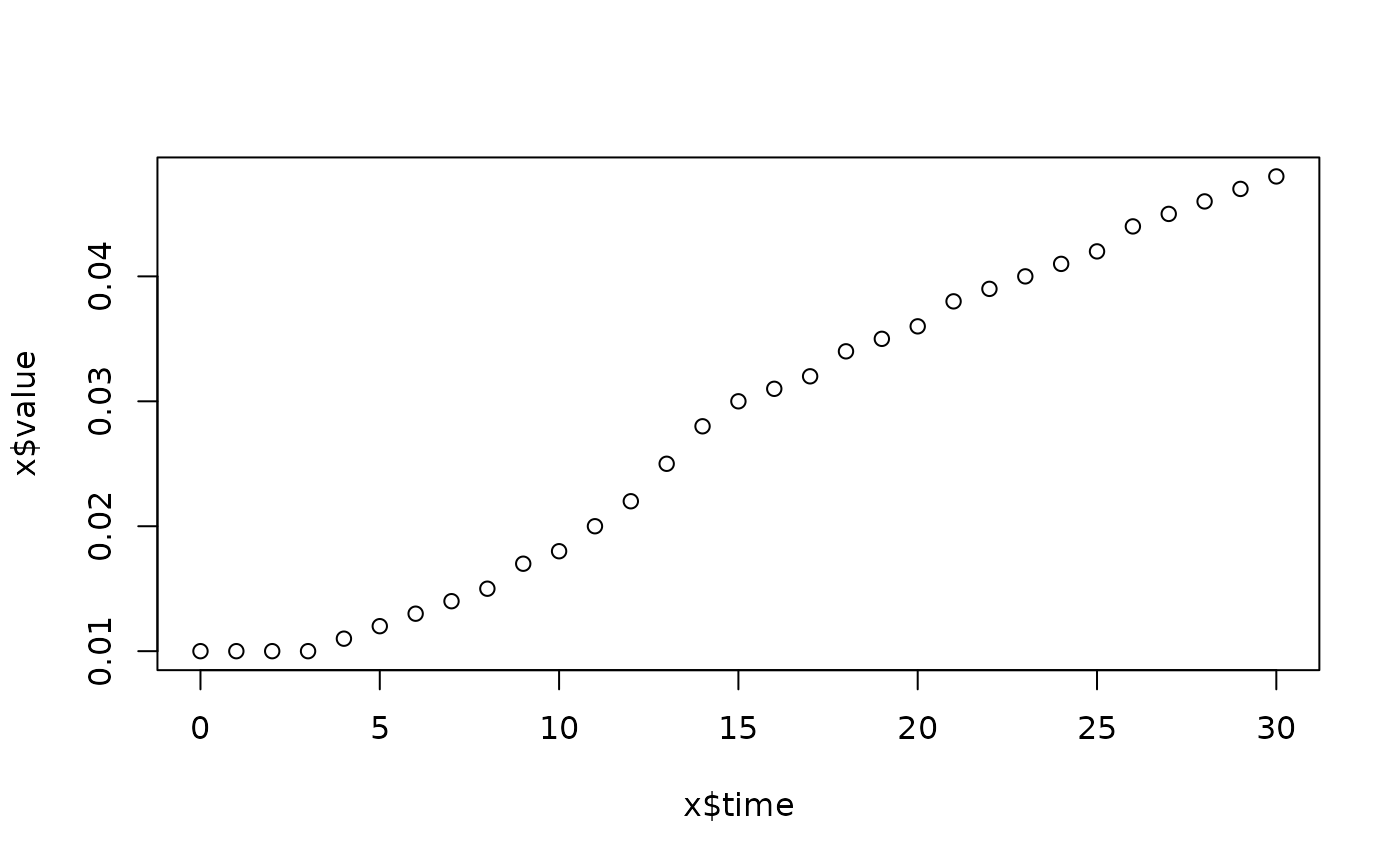
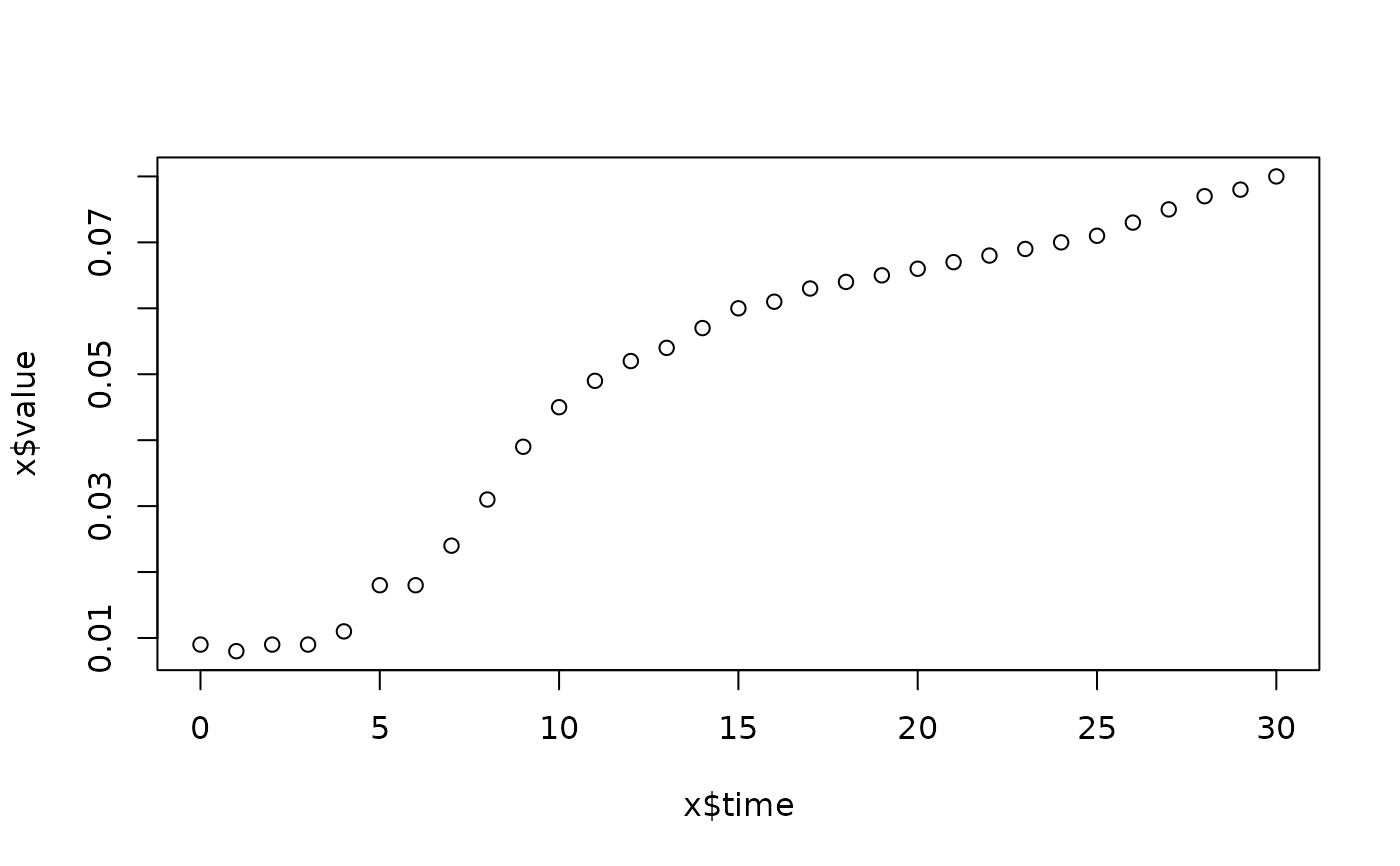
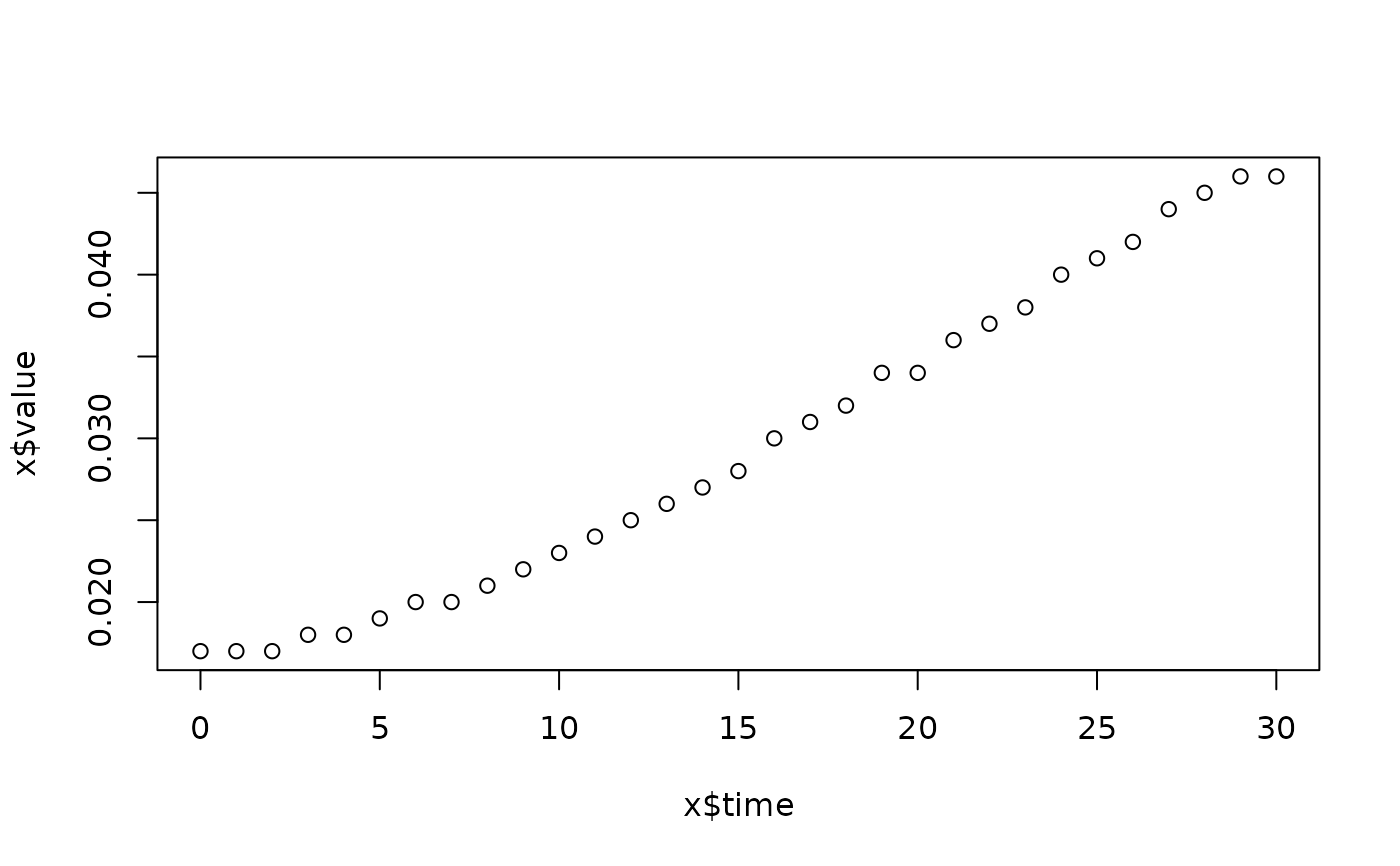
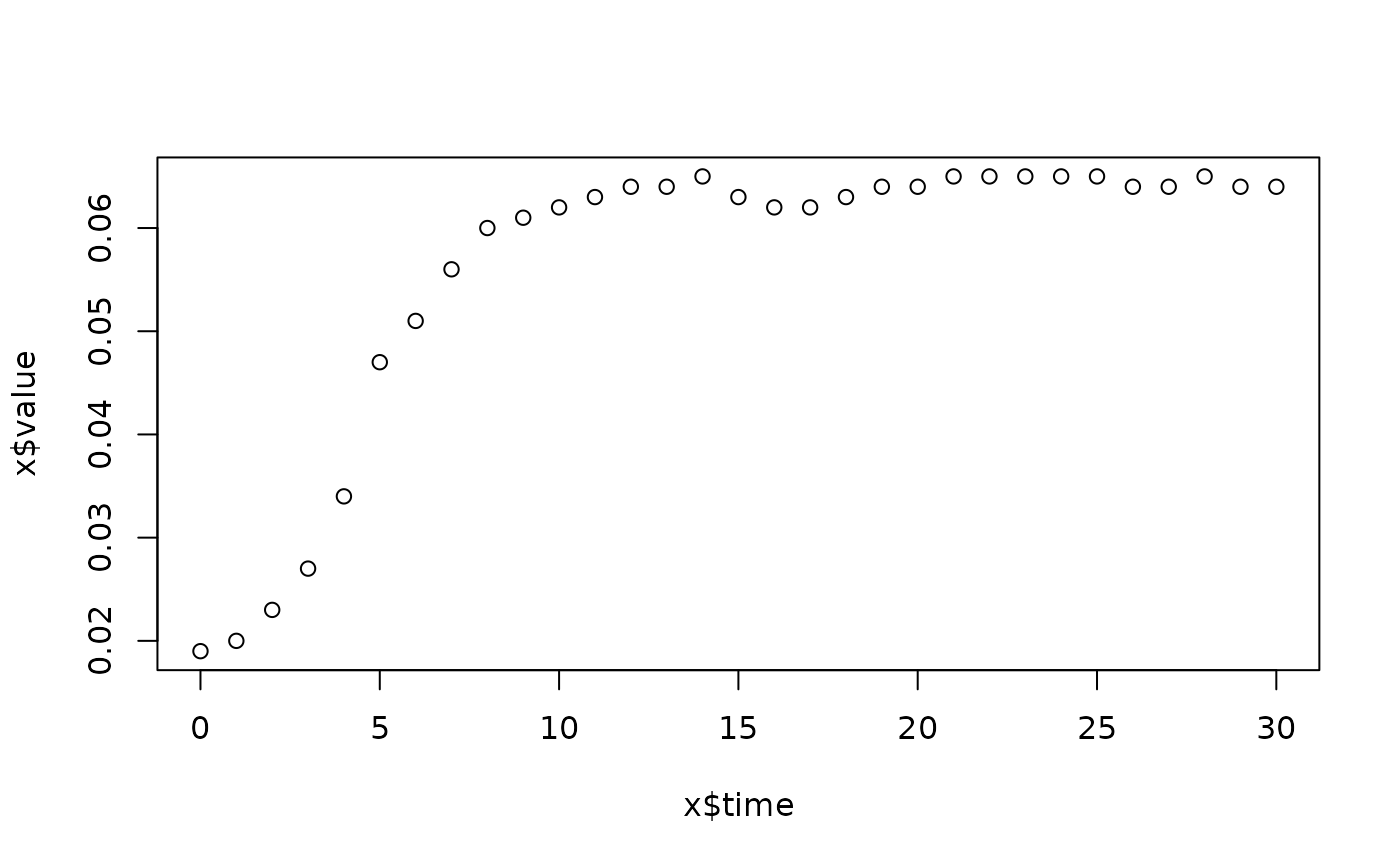
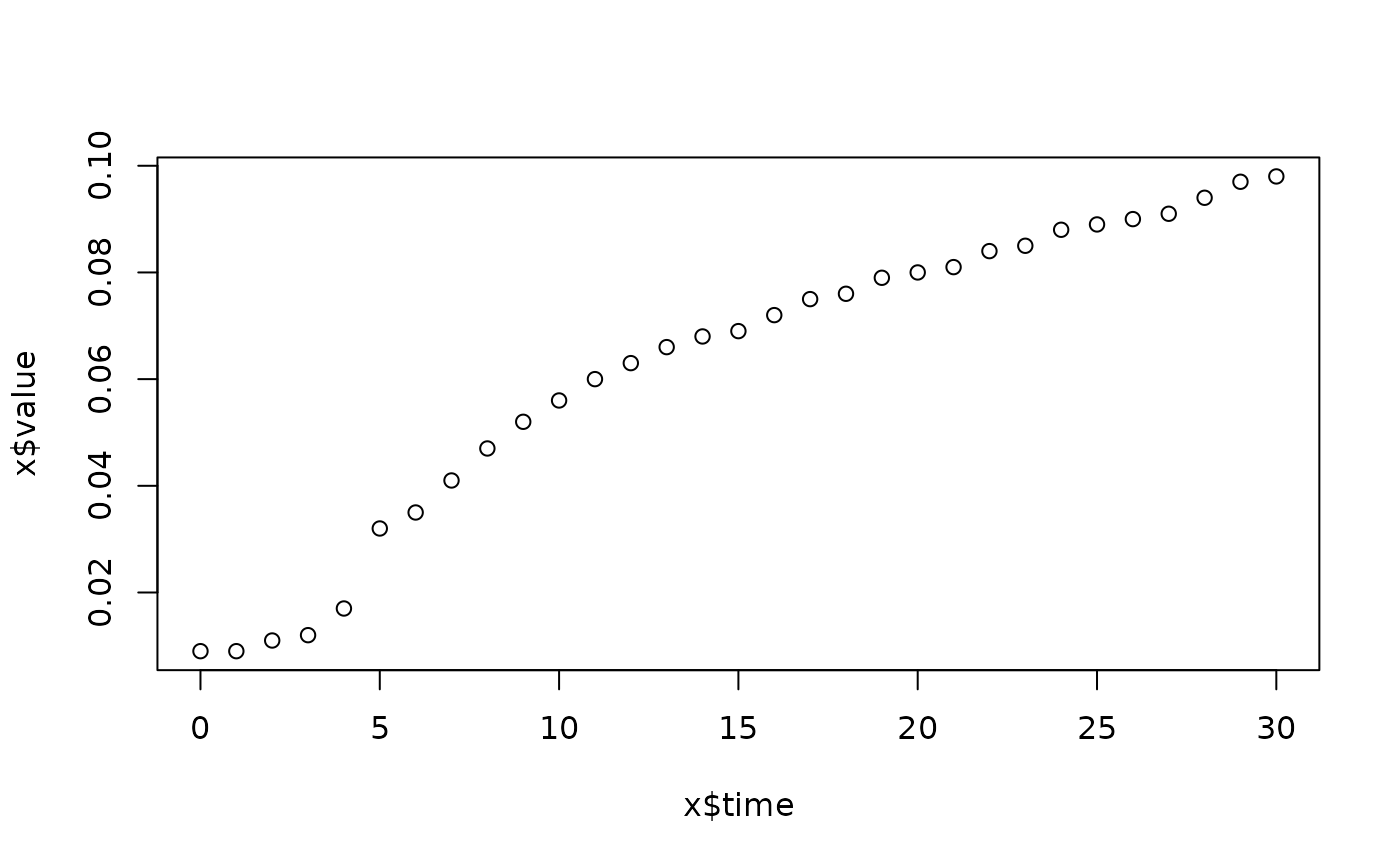
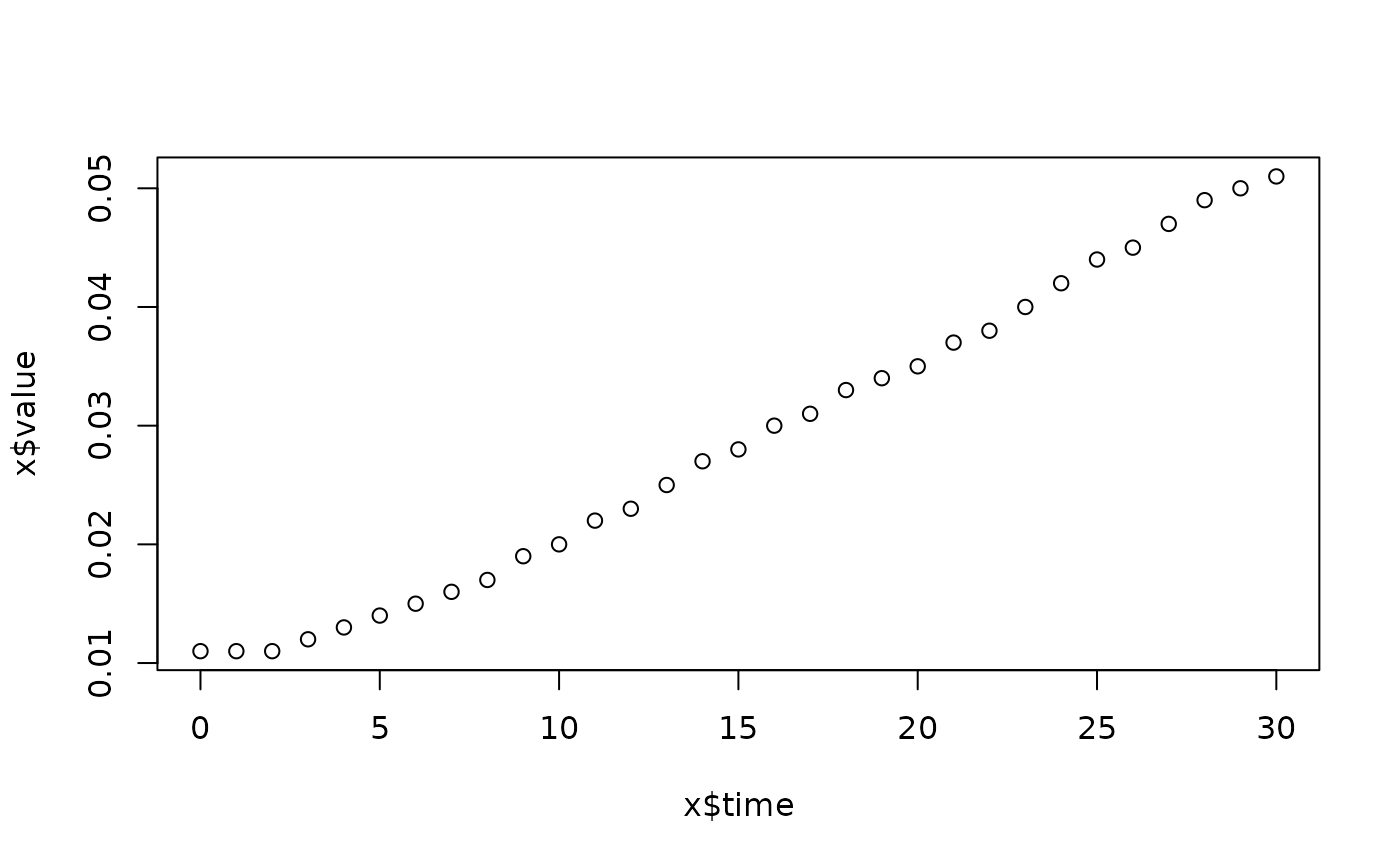
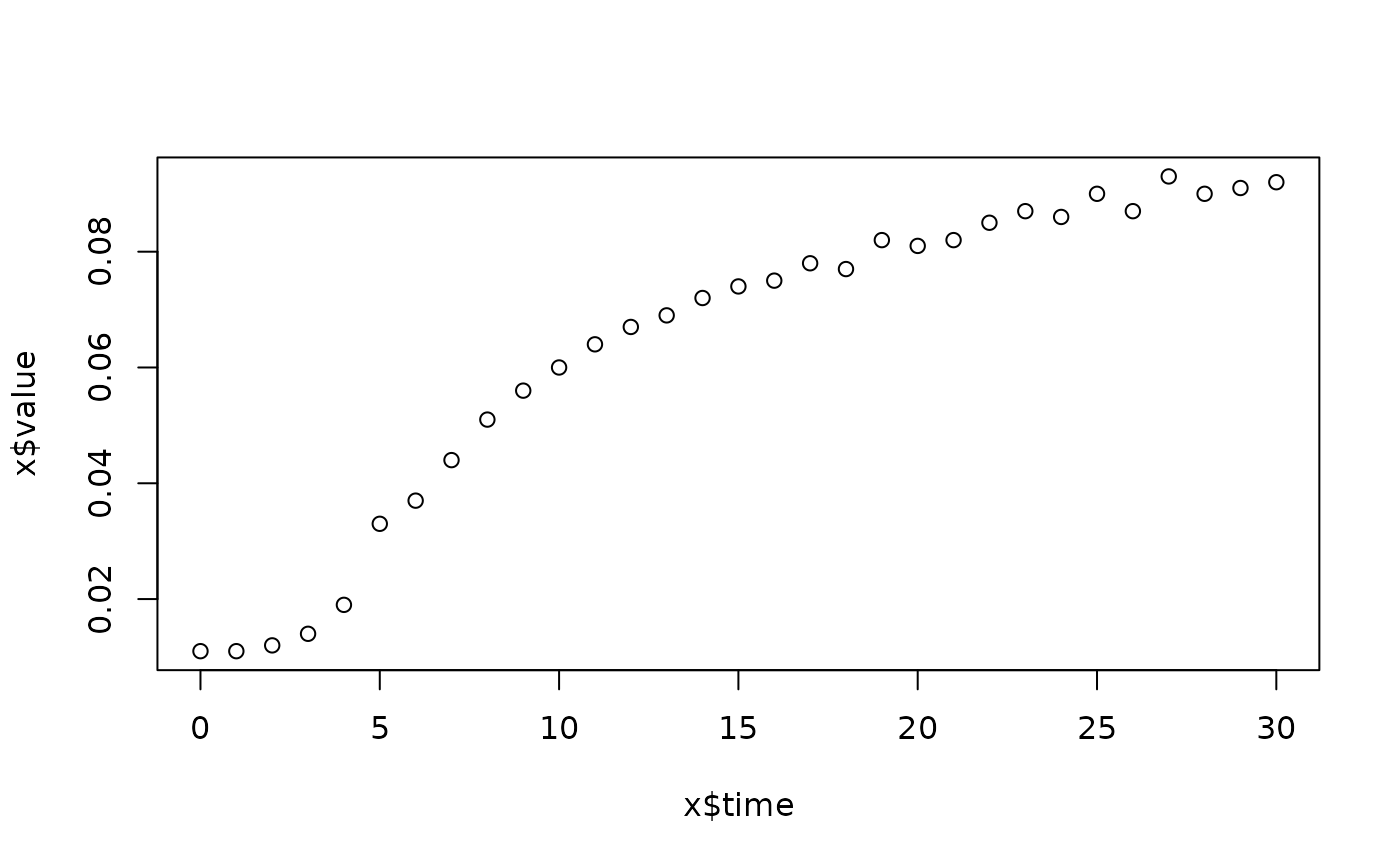
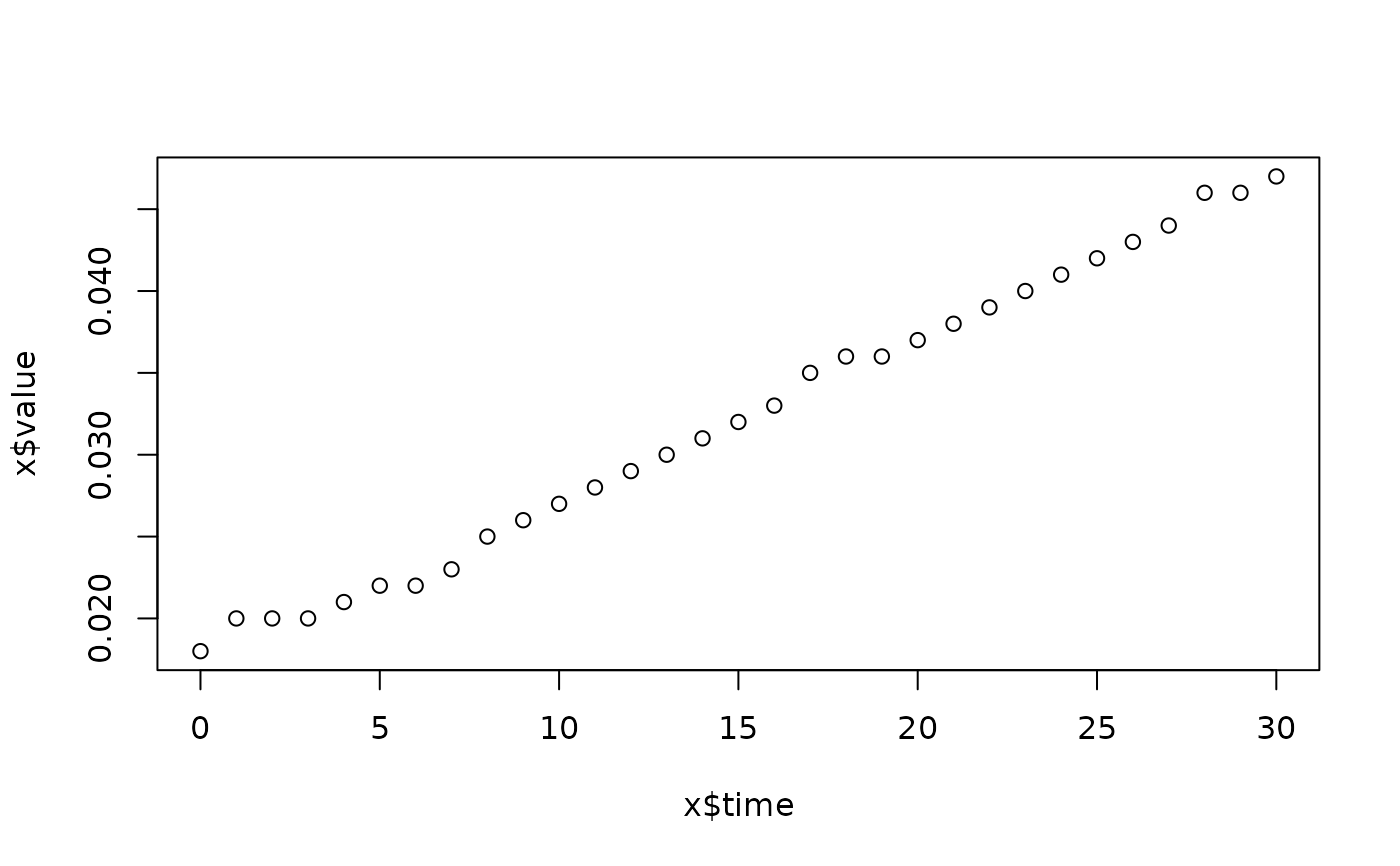
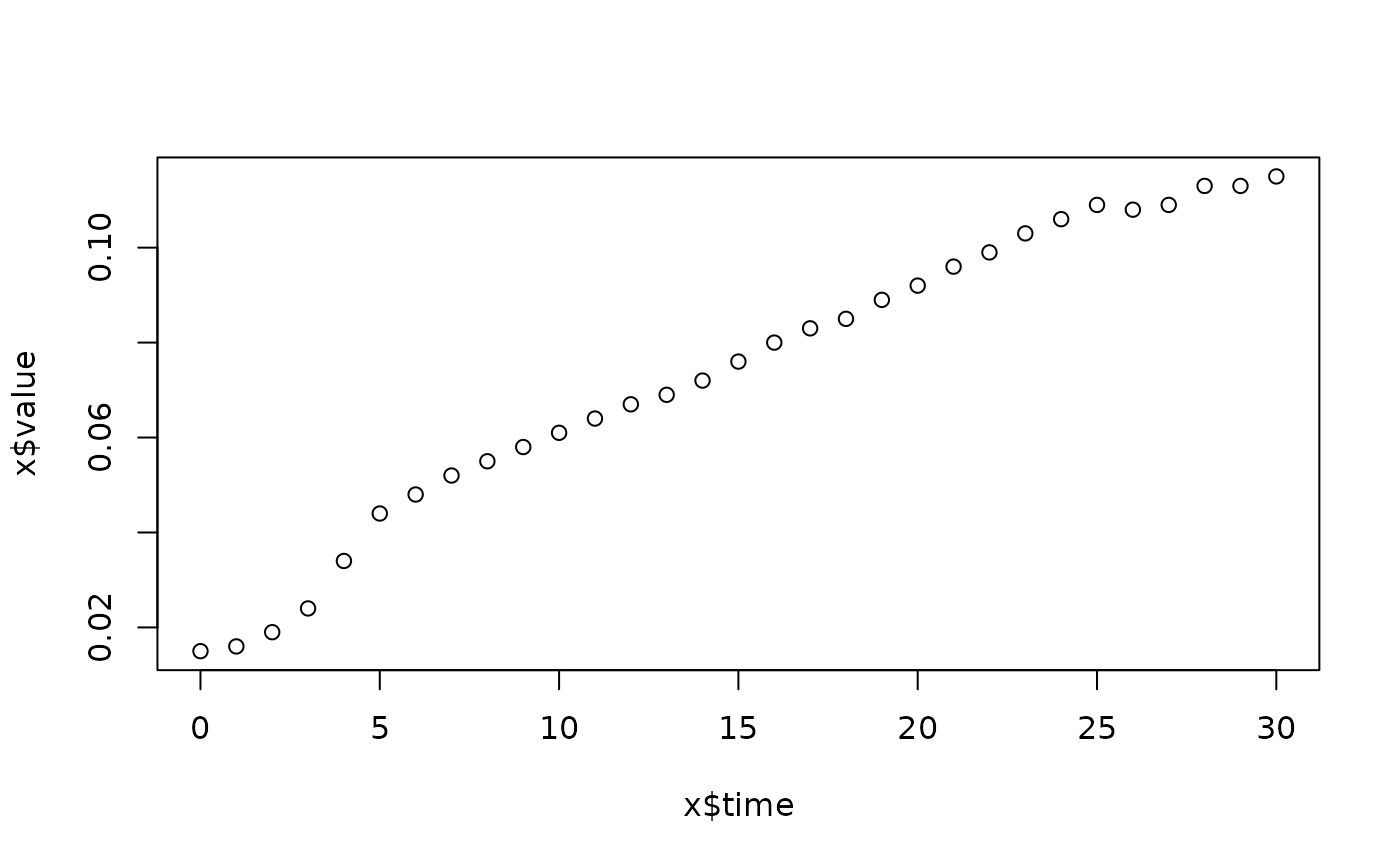
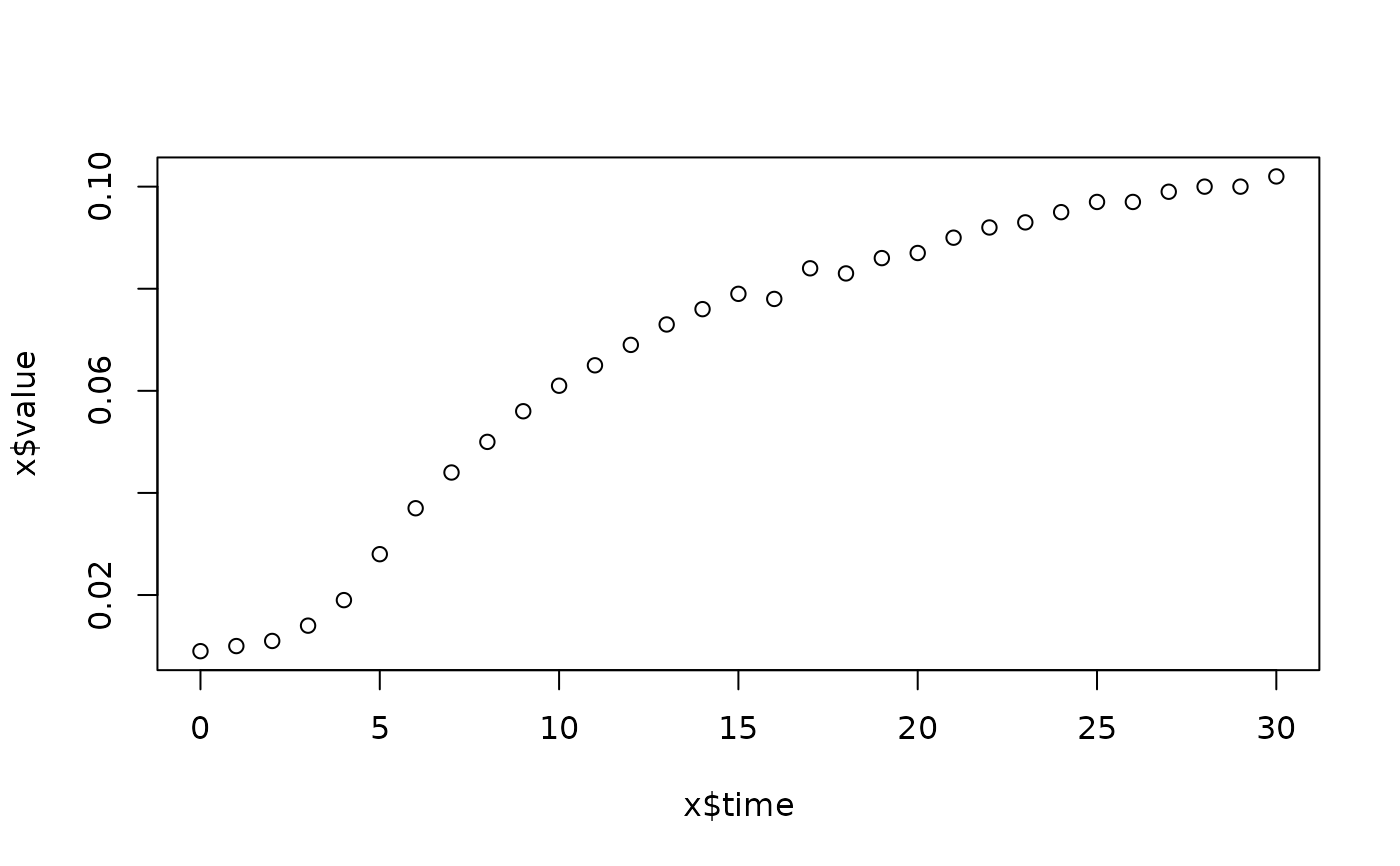
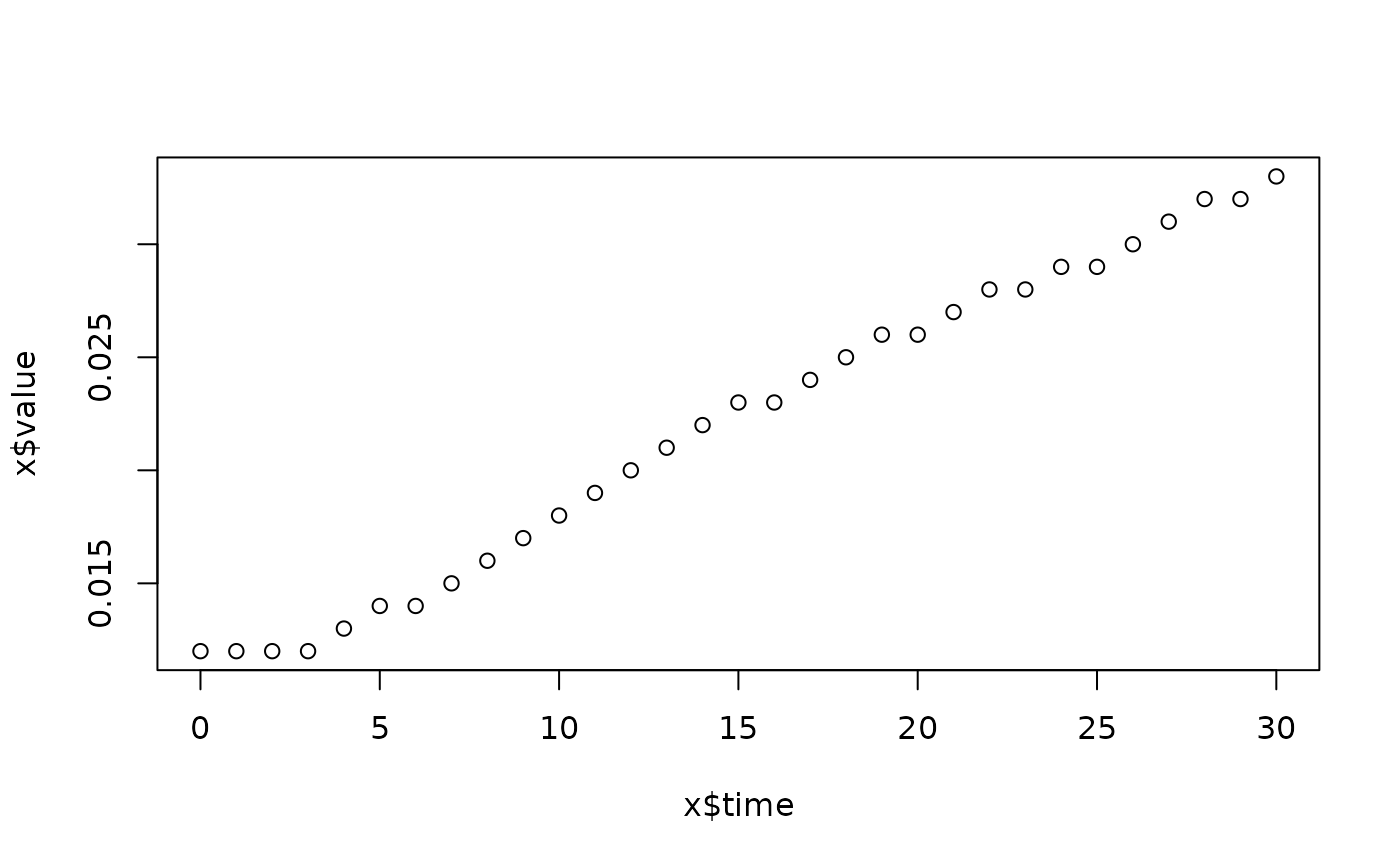
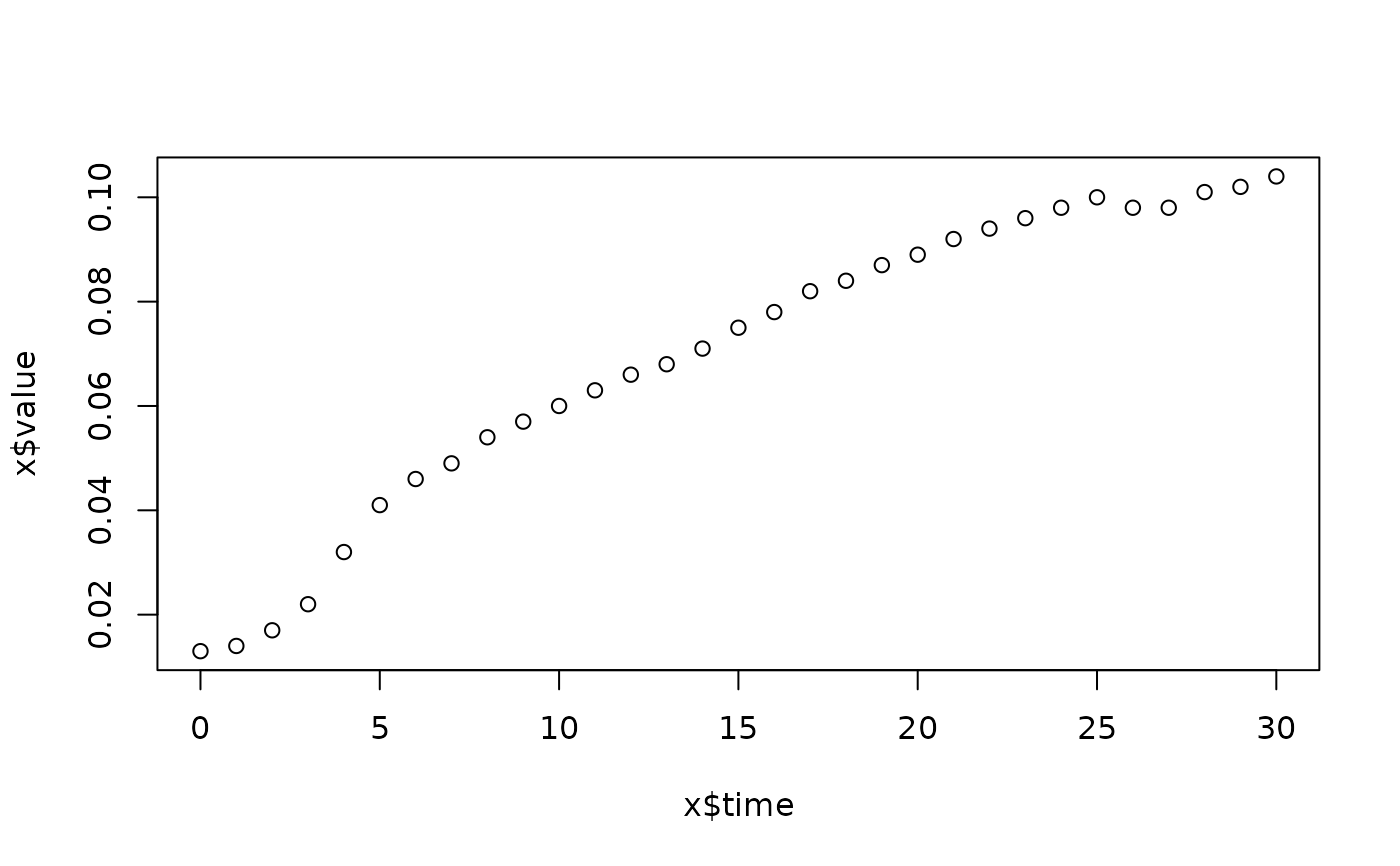
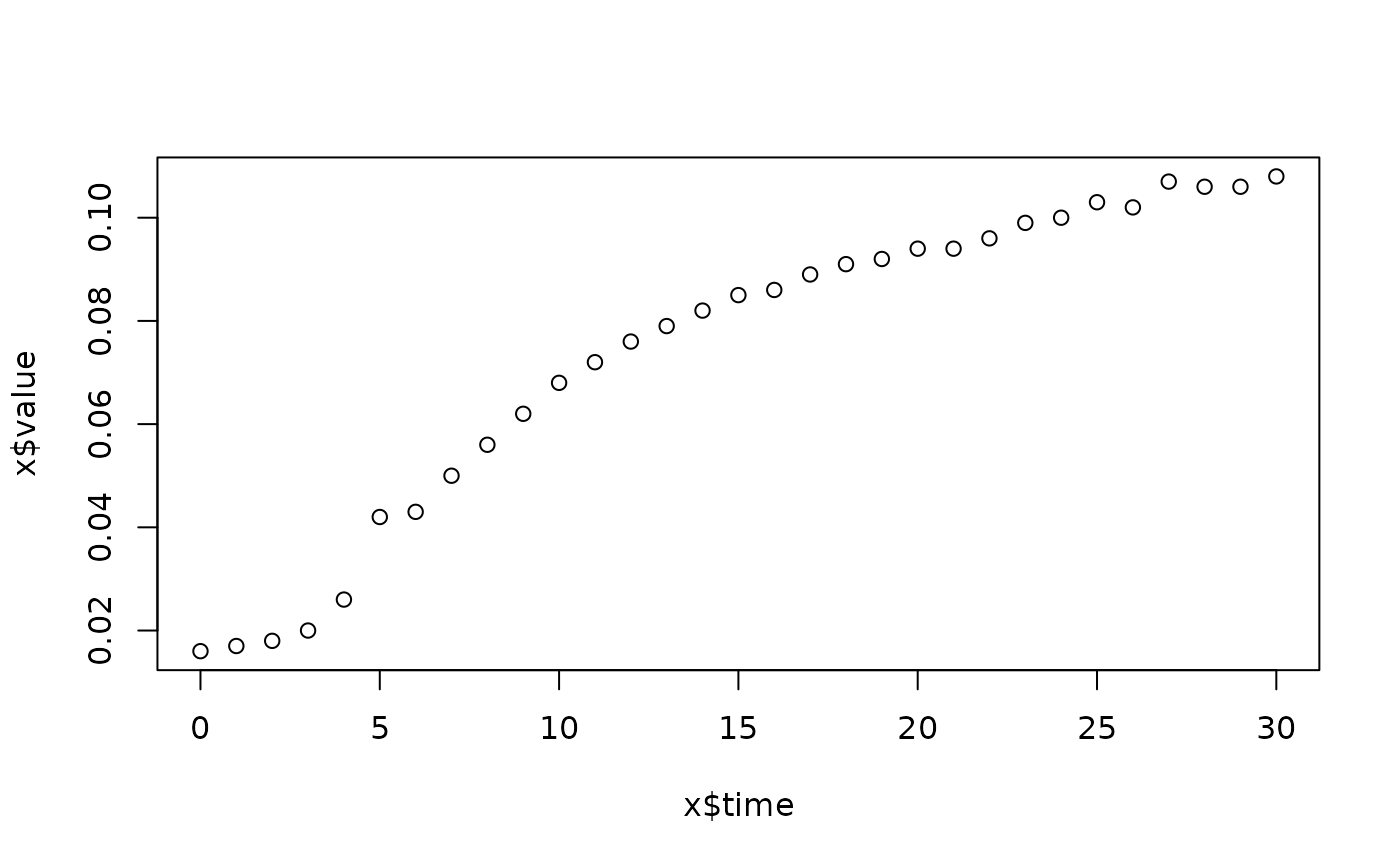
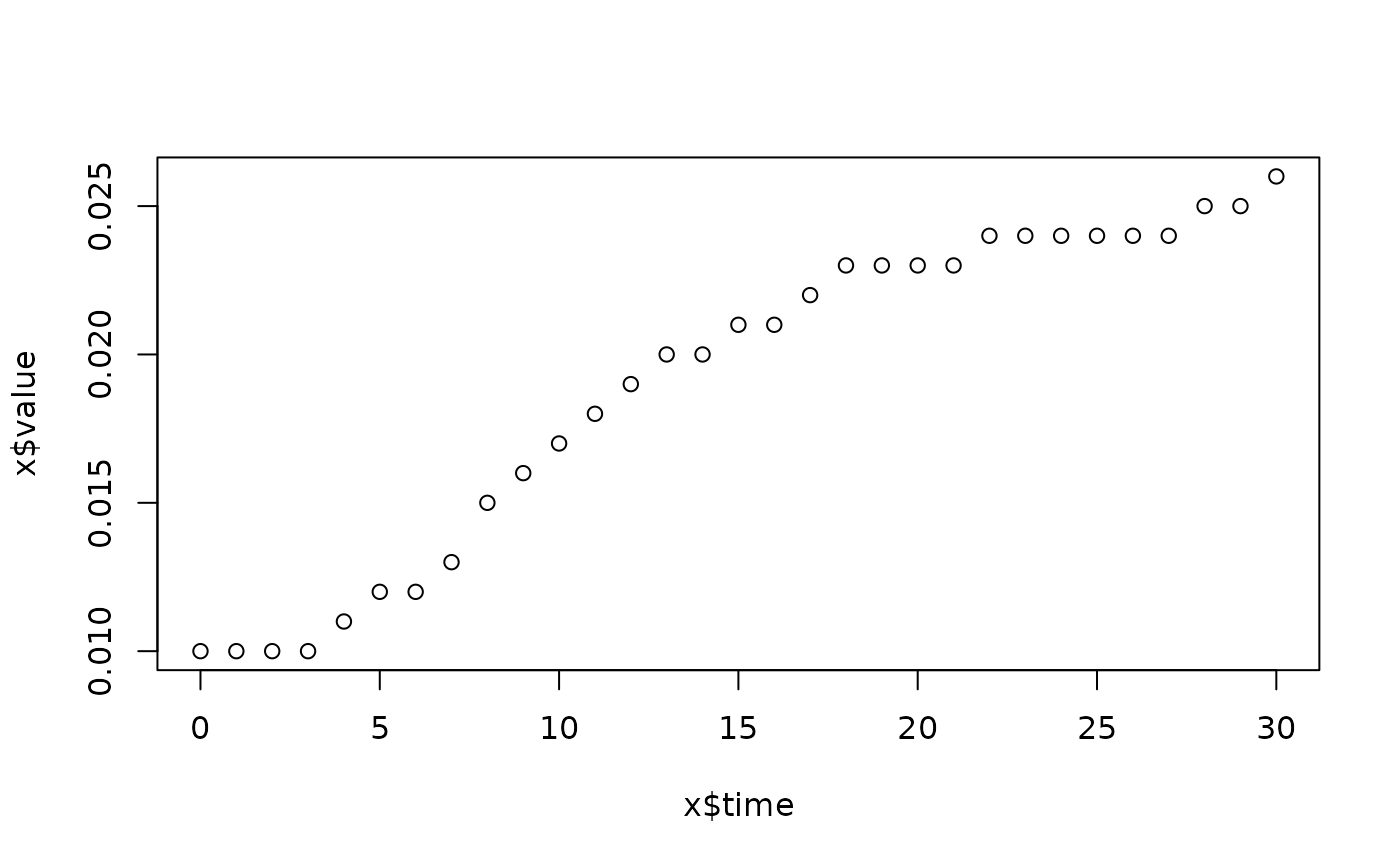
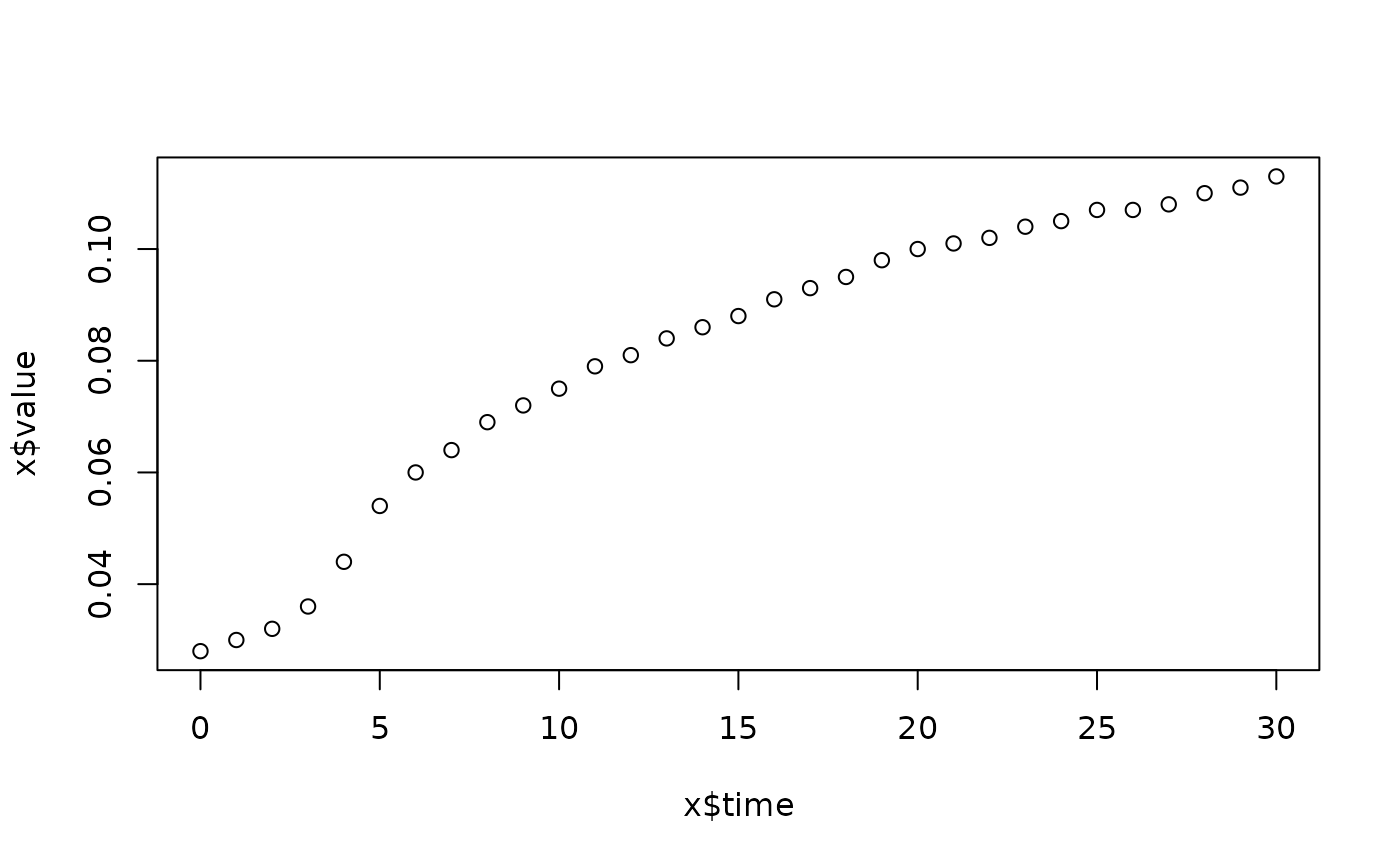
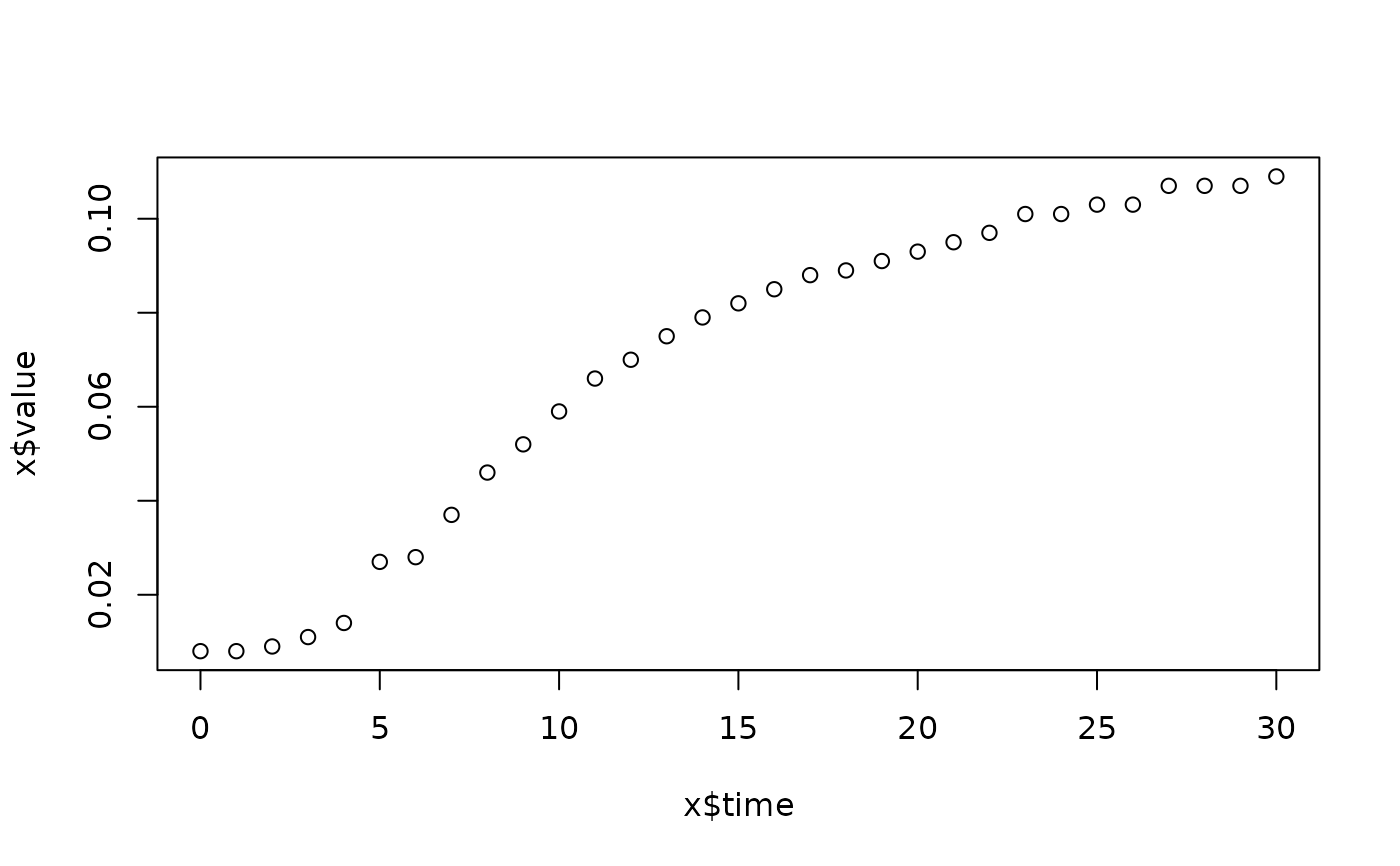
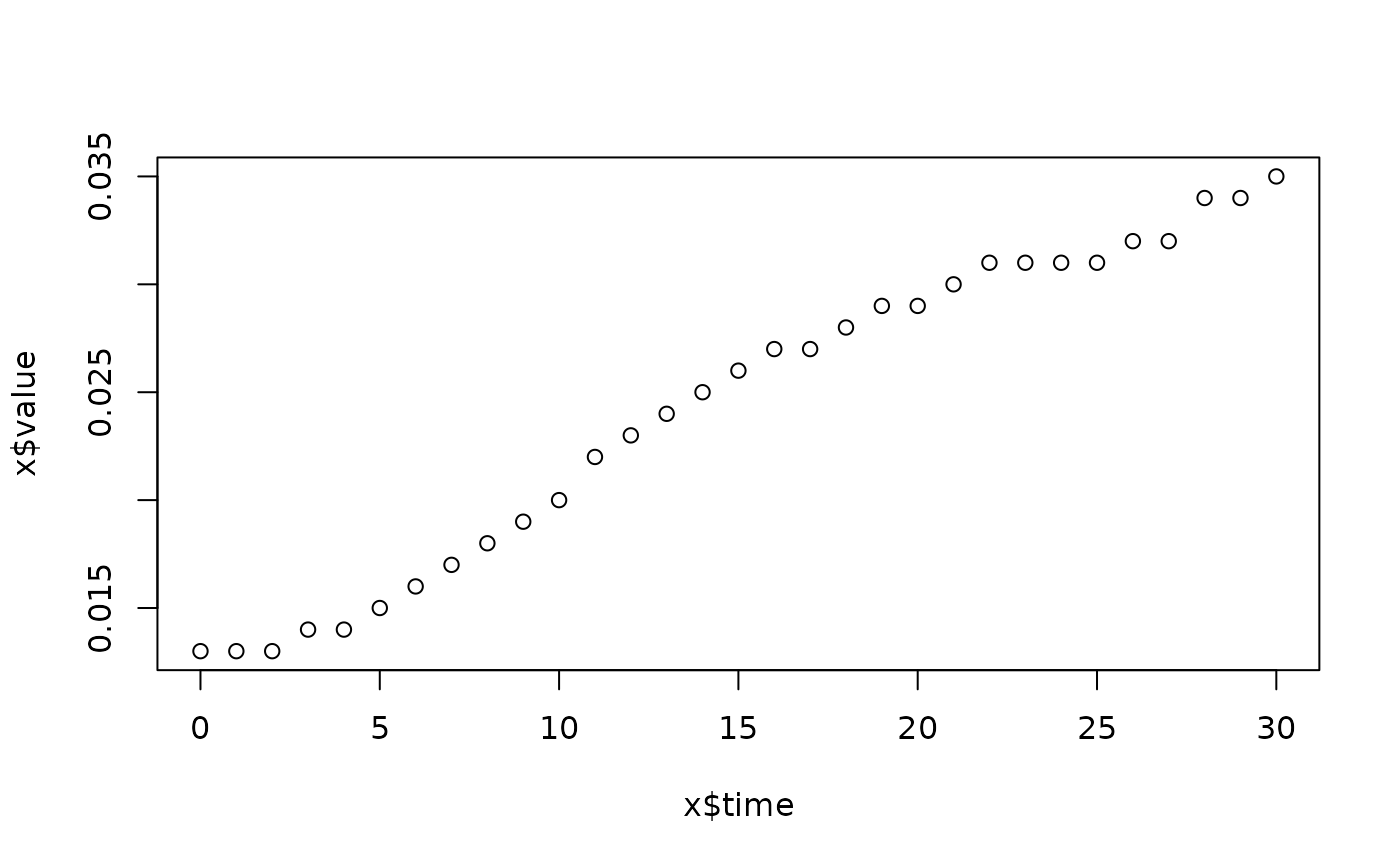
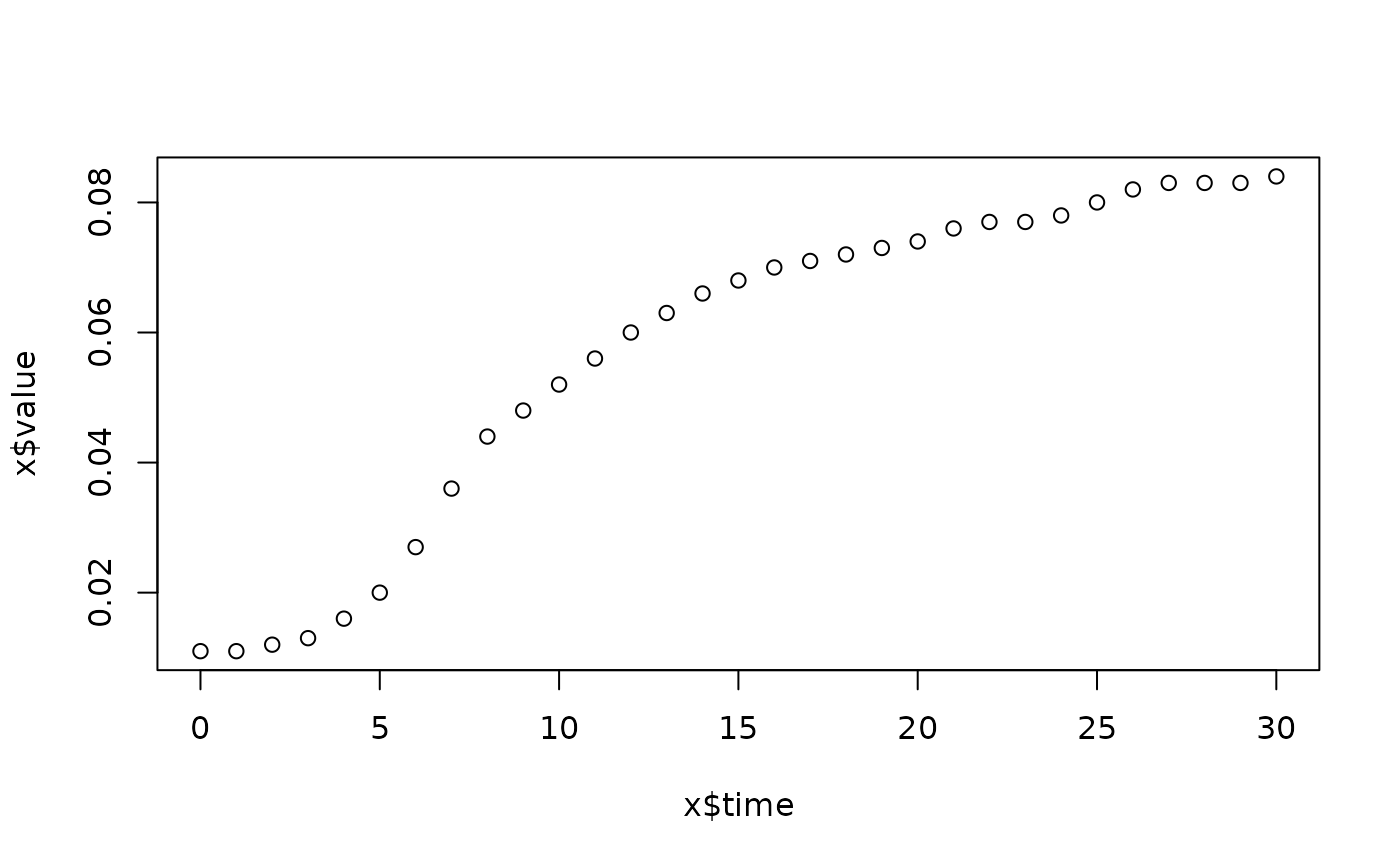
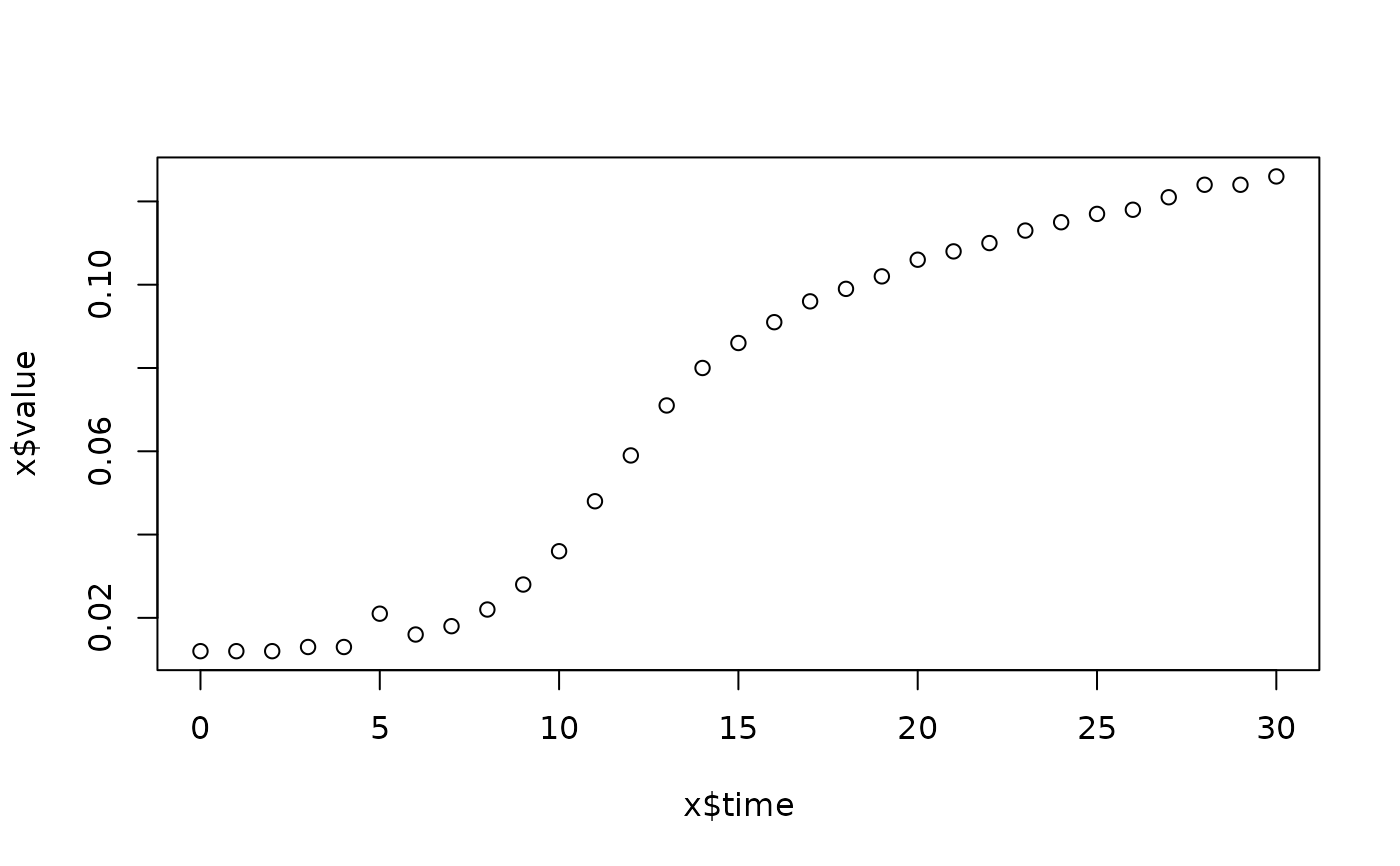
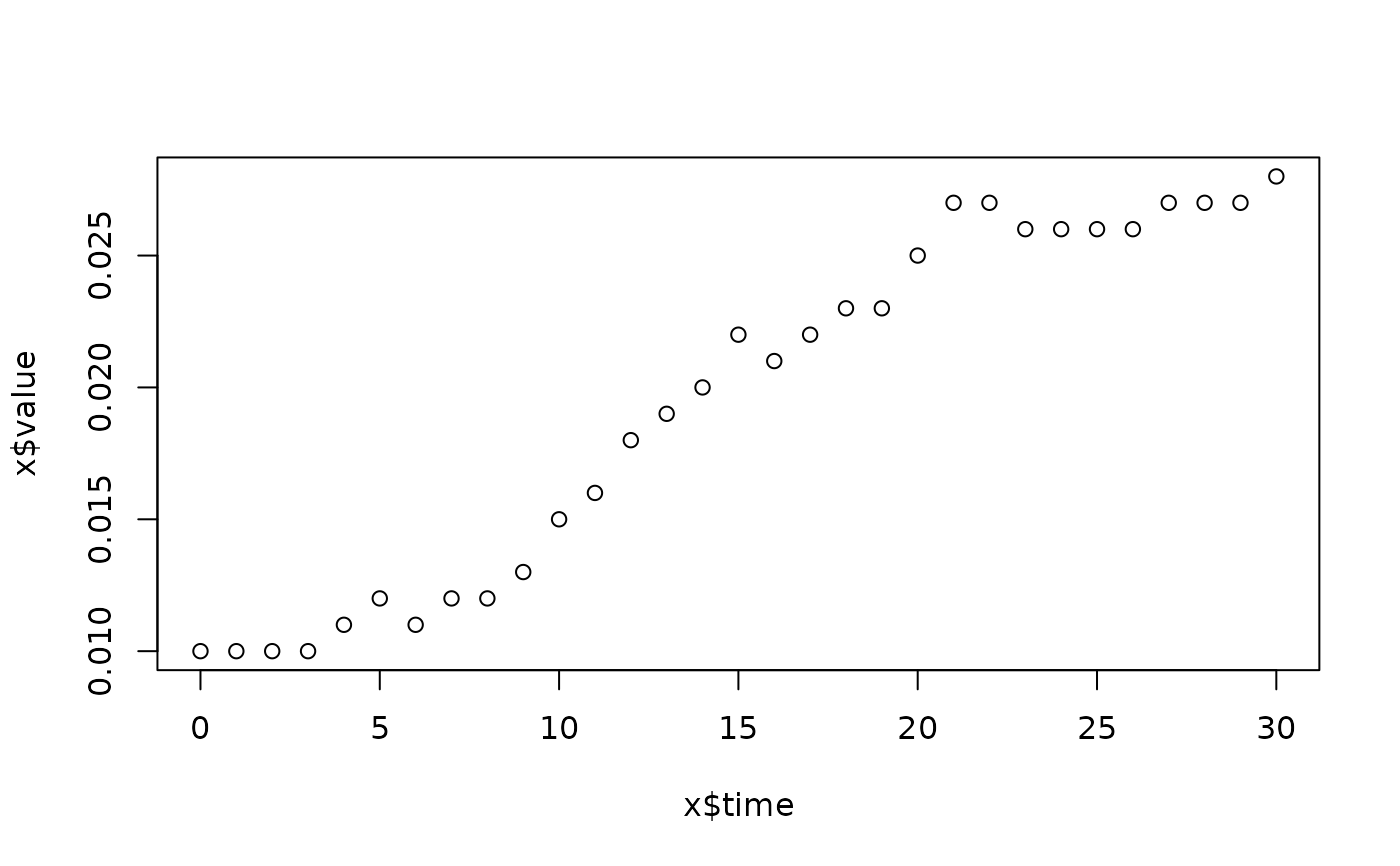
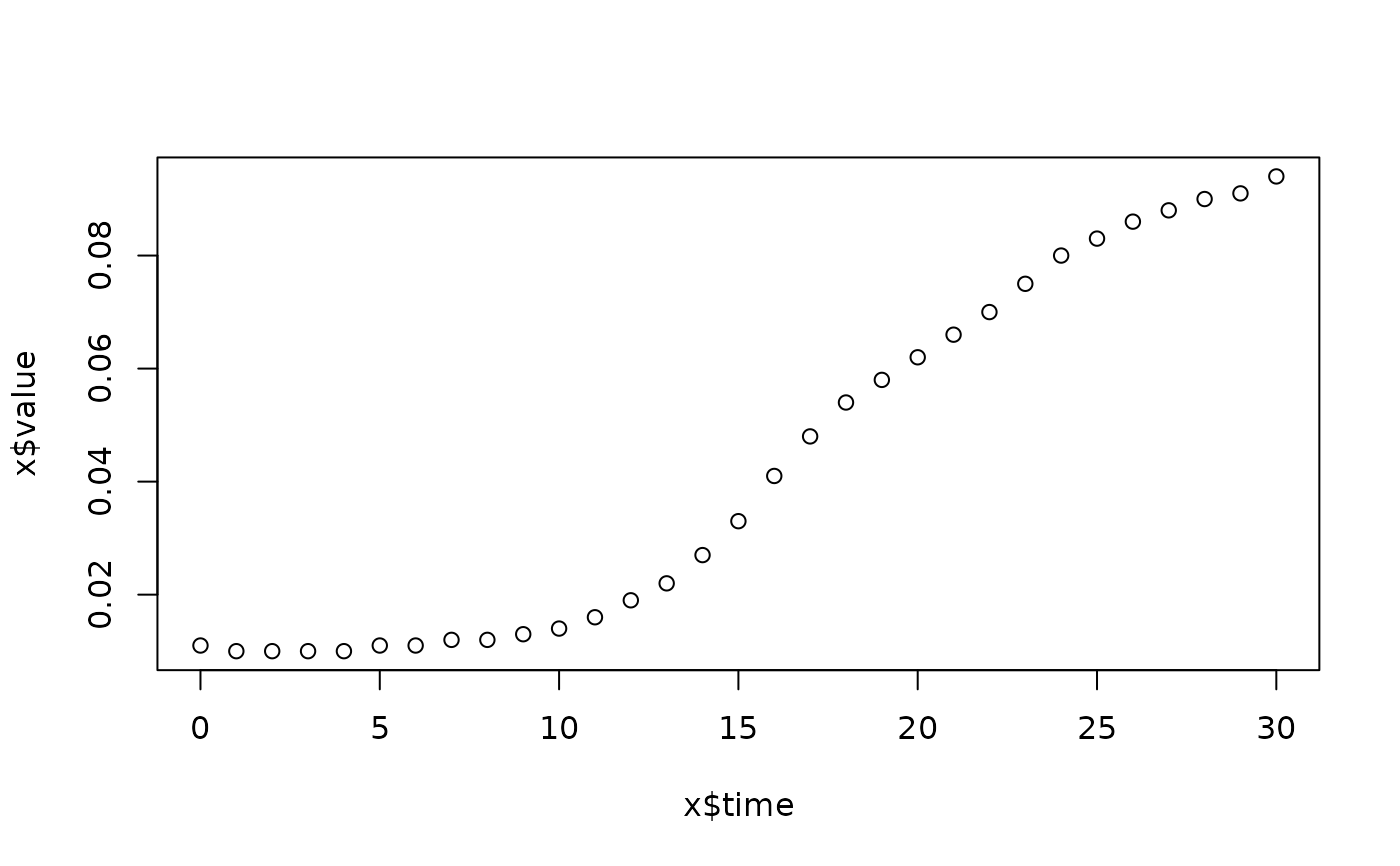
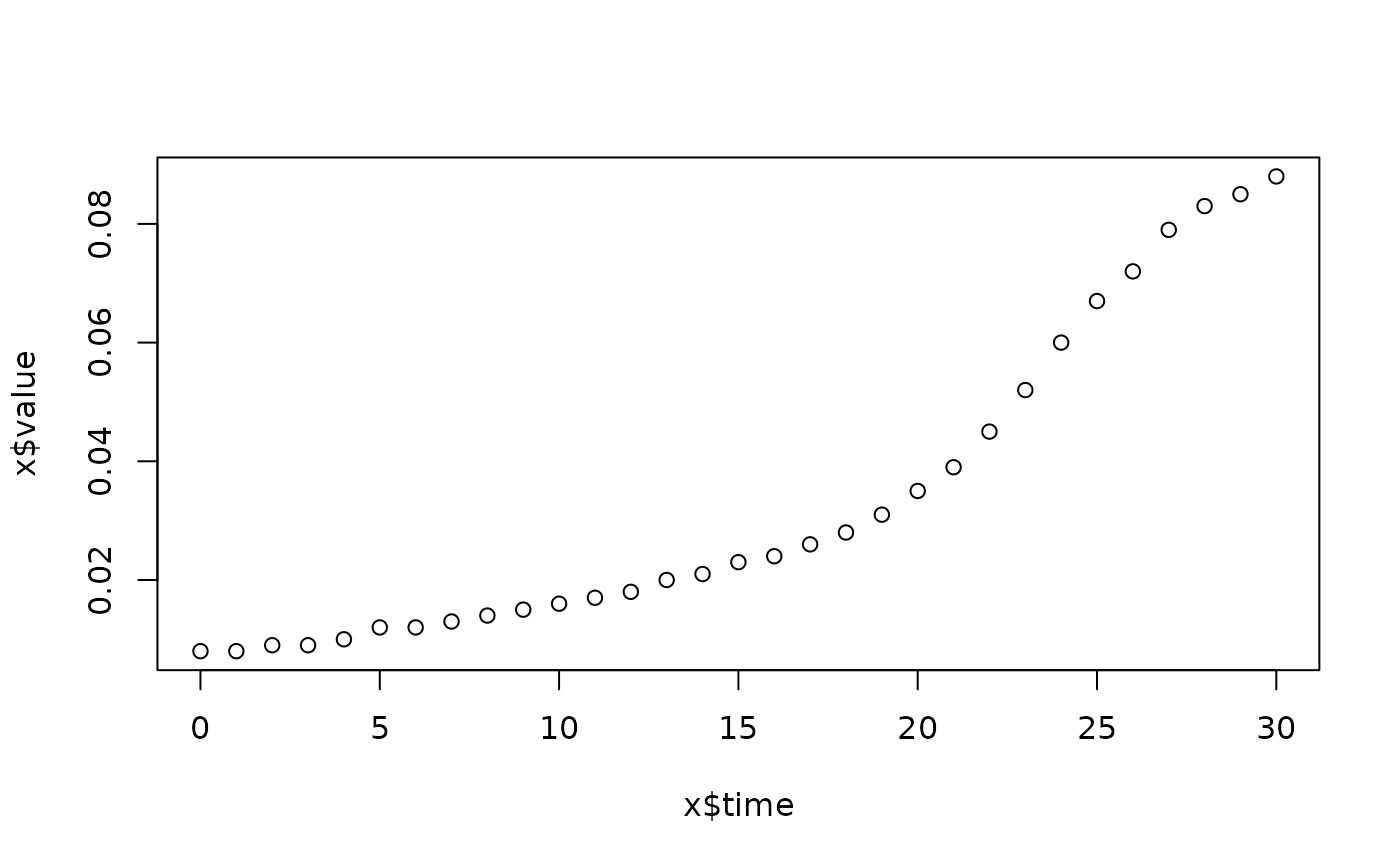
lapply(spl, FUN=function(x)
plot(x$time, x$value,
main=paste(x[1, "strain"], x[1, "conc"], x[1, "replicate"], sep=":")))
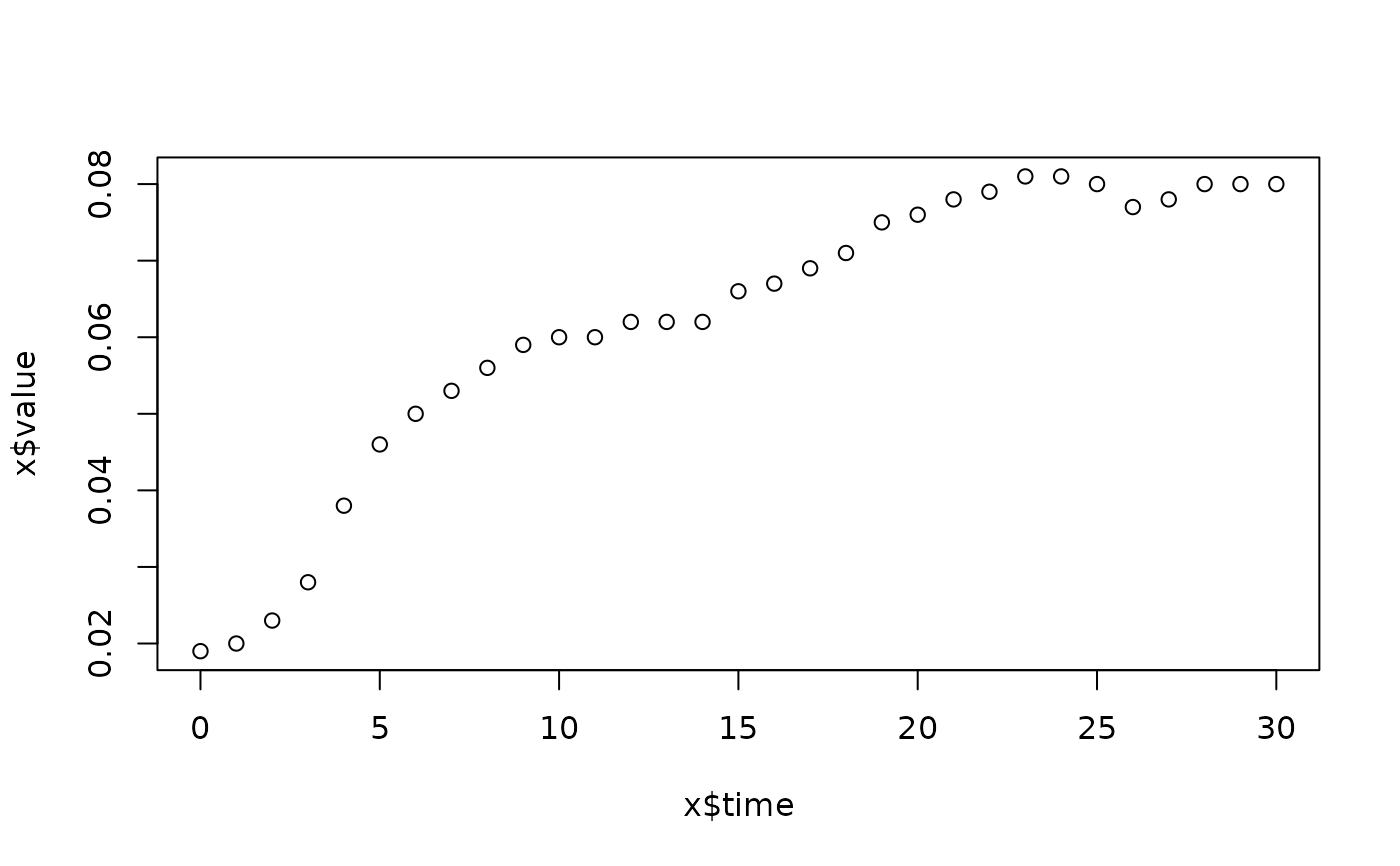
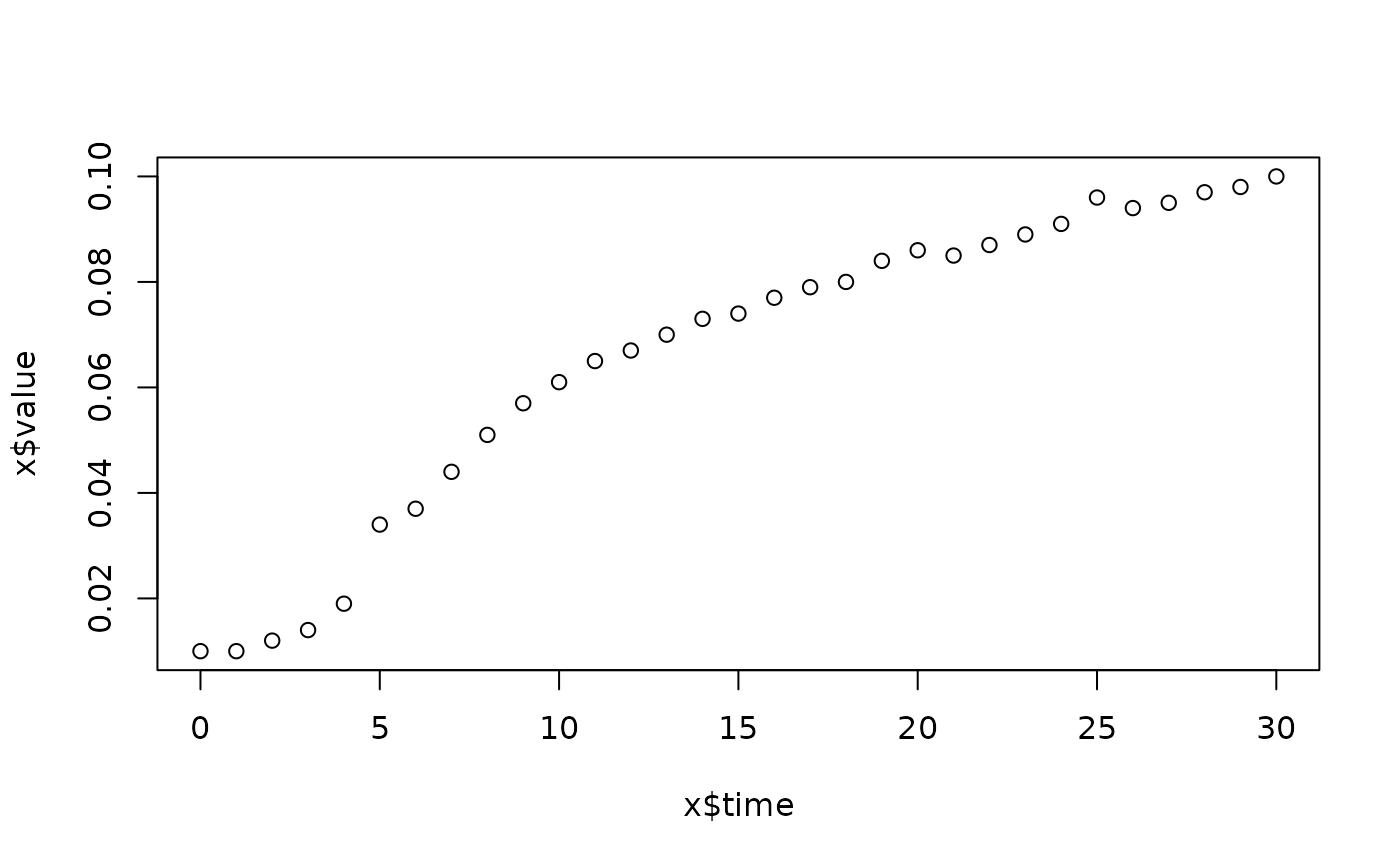
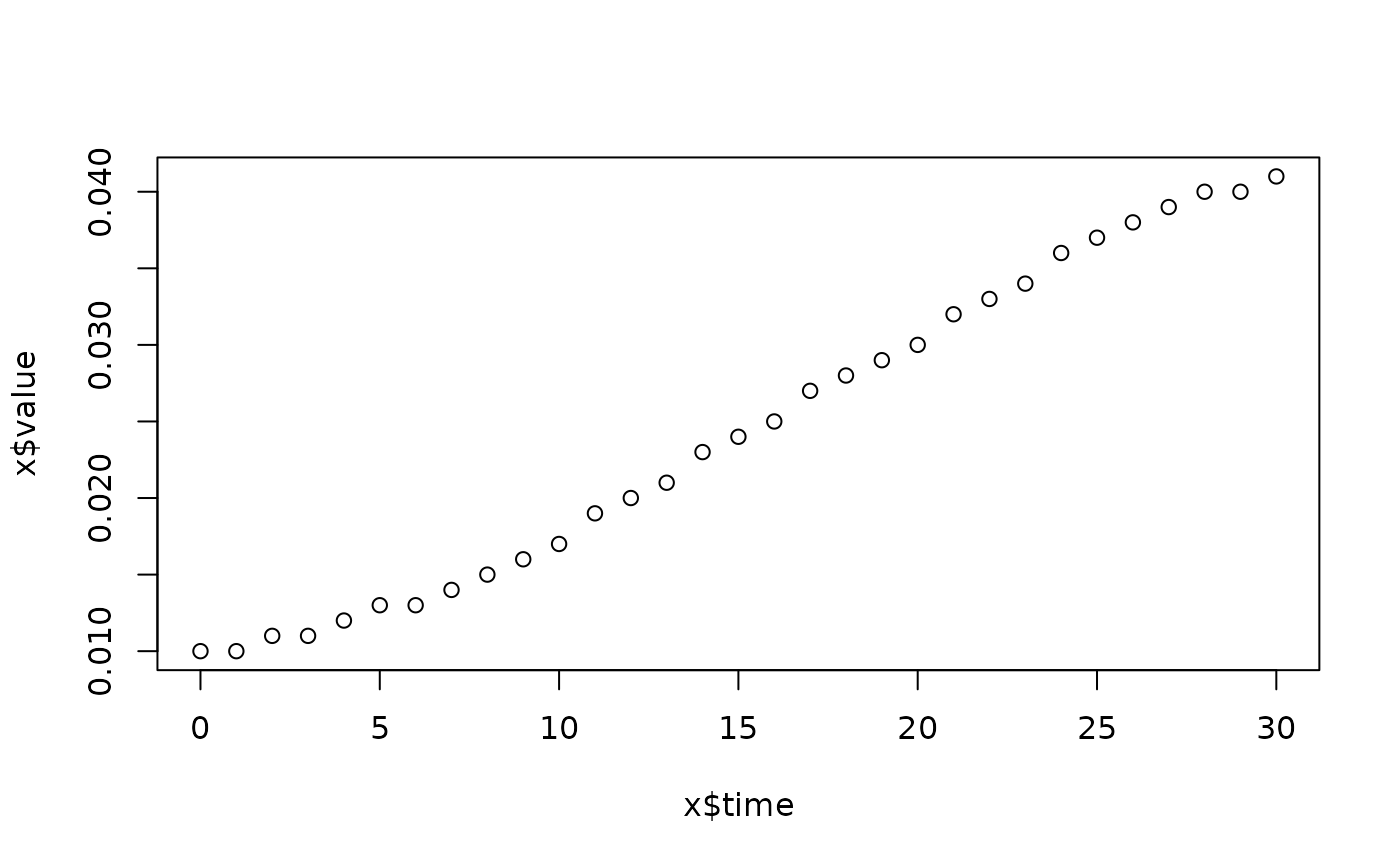
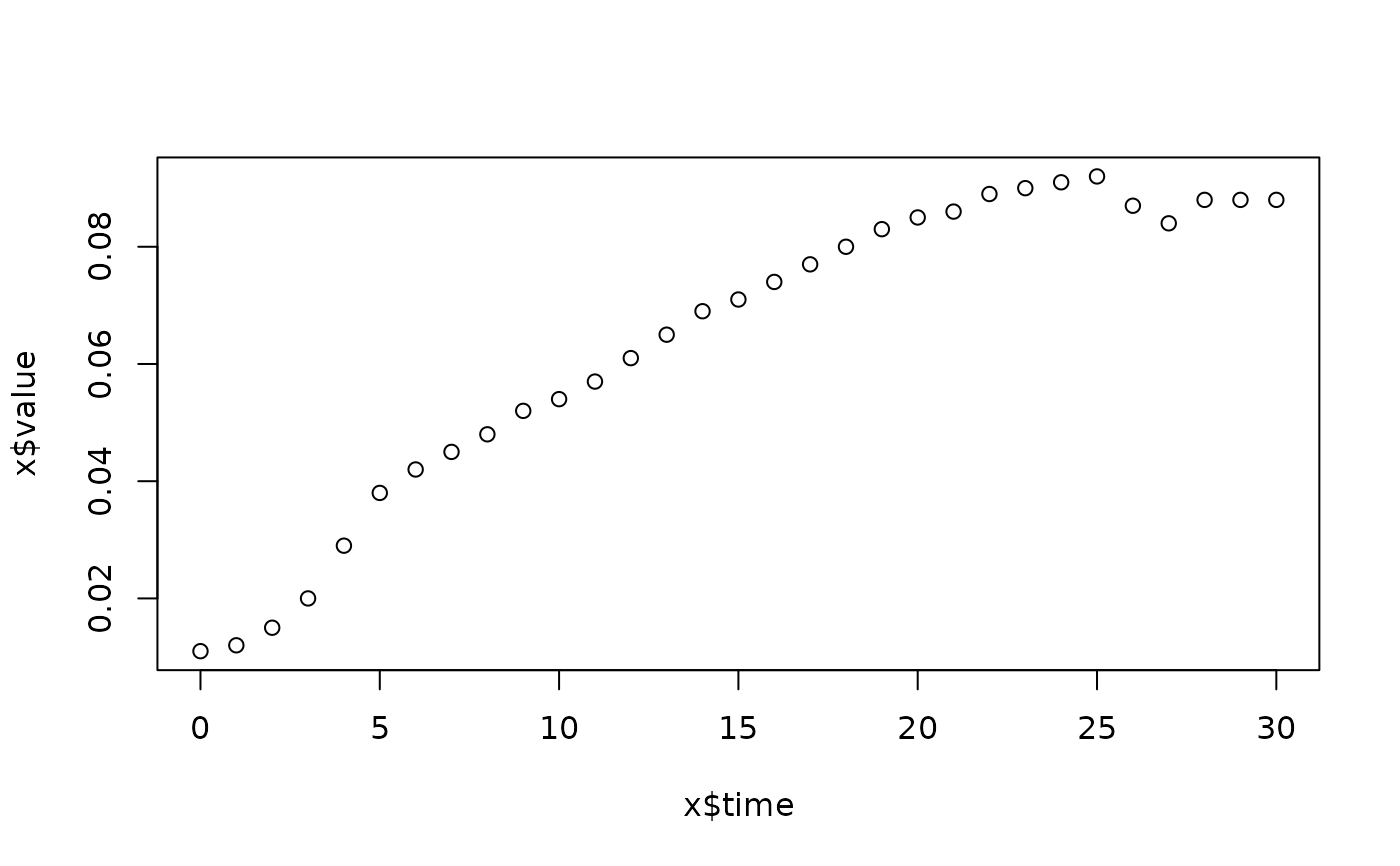
 #> $`D:0:1`
#> NULL
#>
#> $`R:0:1`
#> NULL
#>
#> $`T:0:1`
#> NULL
#>
#> $`D:0.24:1`
#> NULL
#>
#> $`R:0.24:1`
#> NULL
#>
#> $`T:0.24:1`
#> NULL
#>
#> $`D:0.49:1`
#> NULL
#>
#> $`R:0.49:1`
#> NULL
#>
#> $`T:0.49:1`
#> NULL
#>
#> $`D:0.98:1`
#> NULL
#>
#> $`R:0.98:1`
#> NULL
#>
#> $`T:0.98:1`
#> NULL
#>
#> $`D:1.95:1`
#> NULL
#>
#> $`R:1.95:1`
#> NULL
#>
#> $`T:1.95:1`
#> NULL
#>
#> $`D:3.91:1`
#> NULL
#>
#> $`R:3.91:1`
#> NULL
#>
#> $`T:3.91:1`
#> NULL
#>
#> $`D:7.81:1`
#> NULL
#>
#> $`R:7.81:1`
#> NULL
#>
#> $`T:7.81:1`
#> NULL
#>
#> $`D:15.63:1`
#> NULL
#>
#> $`R:15.63:1`
#> NULL
#>
#> $`T:15.63:1`
#> NULL
#>
#> $`D:31.25:1`
#> NULL
#>
#> $`R:31.25:1`
#> NULL
#>
#> $`T:31.25:1`
#> NULL
#>
#> $`D:62.5:1`
#> NULL
#>
#> $`R:62.5:1`
#> NULL
#>
#> $`T:62.5:1`
#> NULL
#>
#> $`D:125:1`
#> NULL
#>
#> $`R:125:1`
#> NULL
#>
#> $`T:125:1`
#> NULL
#>
#> $`D:250:1`
#> NULL
#>
#> $`R:250:1`
#> NULL
#>
#> $`T:250:1`
#> NULL
#>
#> $`D:0:2`
#> NULL
#>
#> $`R:0:2`
#> NULL
#>
#> $`T:0:2`
#> NULL
#>
#> $`D:0.24:2`
#> NULL
#>
#> $`R:0.24:2`
#> NULL
#>
#> $`T:0.24:2`
#> NULL
#>
#> $`D:0.49:2`
#> NULL
#>
#> $`R:0.49:2`
#> NULL
#>
#> $`T:0.49:2`
#> NULL
#>
#> $`D:0.98:2`
#> NULL
#>
#> $`R:0.98:2`
#> NULL
#>
#> $`T:0.98:2`
#> NULL
#>
#> $`D:1.95:2`
#> NULL
#>
#> $`R:1.95:2`
#> NULL
#>
#> $`T:1.95:2`
#> NULL
#>
#> $`D:3.91:2`
#> NULL
#>
#> $`R:3.91:2`
#> NULL
#>
#> $`T:3.91:2`
#> NULL
#>
#> $`D:7.81:2`
#> NULL
#>
#> $`R:7.81:2`
#> NULL
#>
#> $`T:7.81:2`
#> NULL
#>
#> $`D:15.63:2`
#> NULL
#>
#> $`R:15.63:2`
#> NULL
#>
#> $`T:15.63:2`
#> NULL
#>
#> $`D:31.25:2`
#> NULL
#>
#> $`R:31.25:2`
#> NULL
#>
#> $`T:31.25:2`
#> NULL
#>
#> $`D:62.5:2`
#> NULL
#>
#> $`R:62.5:2`
#> NULL
#>
#> $`T:62.5:2`
#> NULL
#>
#> $`D:125:2`
#> NULL
#>
#> $`R:125:2`
#> NULL
#>
#> $`T:125:2`
#> NULL
#>
#> $`D:250:2`
#> NULL
#>
#> $`R:250:2`
#> NULL
#>
#> $`T:250:2`
#> NULL
#>
#> $`D:0:1`
#> NULL
#>
#> $`R:0:1`
#> NULL
#>
#> $`T:0:1`
#> NULL
#>
#> $`D:0.24:1`
#> NULL
#>
#> $`R:0.24:1`
#> NULL
#>
#> $`T:0.24:1`
#> NULL
#>
#> $`D:0.49:1`
#> NULL
#>
#> $`R:0.49:1`
#> NULL
#>
#> $`T:0.49:1`
#> NULL
#>
#> $`D:0.98:1`
#> NULL
#>
#> $`R:0.98:1`
#> NULL
#>
#> $`T:0.98:1`
#> NULL
#>
#> $`D:1.95:1`
#> NULL
#>
#> $`R:1.95:1`
#> NULL
#>
#> $`T:1.95:1`
#> NULL
#>
#> $`D:3.91:1`
#> NULL
#>
#> $`R:3.91:1`
#> NULL
#>
#> $`T:3.91:1`
#> NULL
#>
#> $`D:7.81:1`
#> NULL
#>
#> $`R:7.81:1`
#> NULL
#>
#> $`T:7.81:1`
#> NULL
#>
#> $`D:15.63:1`
#> NULL
#>
#> $`R:15.63:1`
#> NULL
#>
#> $`T:15.63:1`
#> NULL
#>
#> $`D:31.25:1`
#> NULL
#>
#> $`R:31.25:1`
#> NULL
#>
#> $`T:31.25:1`
#> NULL
#>
#> $`D:62.5:1`
#> NULL
#>
#> $`R:62.5:1`
#> NULL
#>
#> $`T:62.5:1`
#> NULL
#>
#> $`D:125:1`
#> NULL
#>
#> $`R:125:1`
#> NULL
#>
#> $`T:125:1`
#> NULL
#>
#> $`D:250:1`
#> NULL
#>
#> $`R:250:1`
#> NULL
#>
#> $`T:250:1`
#> NULL
#>
#> $`D:0:2`
#> NULL
#>
#> $`R:0:2`
#> NULL
#>
#> $`T:0:2`
#> NULL
#>
#> $`D:0.24:2`
#> NULL
#>
#> $`R:0.24:2`
#> NULL
#>
#> $`T:0.24:2`
#> NULL
#>
#> $`D:0.49:2`
#> NULL
#>
#> $`R:0.49:2`
#> NULL
#>
#> $`T:0.49:2`
#> NULL
#>
#> $`D:0.98:2`
#> NULL
#>
#> $`R:0.98:2`
#> NULL
#>
#> $`T:0.98:2`
#> NULL
#>
#> $`D:1.95:2`
#> NULL
#>
#> $`R:1.95:2`
#> NULL
#>
#> $`T:1.95:2`
#> NULL
#>
#> $`D:3.91:2`
#> NULL
#>
#> $`R:3.91:2`
#> NULL
#>
#> $`T:3.91:2`
#> NULL
#>
#> $`D:7.81:2`
#> NULL
#>
#> $`R:7.81:2`
#> NULL
#>
#> $`T:7.81:2`
#> NULL
#>
#> $`D:15.63:2`
#> NULL
#>
#> $`R:15.63:2`
#> NULL
#>
#> $`T:15.63:2`
#> NULL
#>
#> $`D:31.25:2`
#> NULL
#>
#> $`R:31.25:2`
#> NULL
#>
#> $`T:31.25:2`
#> NULL
#>
#> $`D:62.5:2`
#> NULL
#>
#> $`R:62.5:2`
#> NULL
#>
#> $`T:62.5:2`
#> NULL
#>
#> $`D:125:2`
#> NULL
#>
#> $`R:125:2`
#> NULL
#>
#> $`T:125:2`
#> NULL
#>
#> $`D:250:2`
#> NULL
#>
#> $`R:250:2`
#> NULL
#>
#> $`T:250:2`
#> NULL
#>