Start Values for ECOFFinder
ecoffinder_startpar.RdA heuristic function functon to estimate start values for the ECOFFinder.
ecoffinder_startpar(x, y, method = c("peak1", "mode", "mean"), bw = "nrd0")
Arguments
| x | x-values (log2 MIC or inhibition zone diameter) |
|---|---|
| y | y-values (frequencies) |
| method | select heuristic "peak1", "mode" or "mean" |
| bw | bandwith of kernel density smoother (see |
Value
vector with estimates for location, scale and total frequency
Details
Start values for the nonlinear regression are estimated from the mode and median absolute deviation, from mean and standard deviation of the data or from a heuristic peak detection method, borrowed from package cardidates.
References
Rolinski, S., Sachse, S. and Petzoldt, T. (2007) cardidates: Identification of Cardinal Dates in Ecological Time Series. R package version 0.4.8. https://cran.r-project.org/package=cardidates
Turnidge, J., Kahlmeter, G., Kronvall, G. (2006) Statistical characterization of bacterial wild-type MIC value distributions and the determination of epidemiological cut-off values. Clin Microbial Infect 12: 418-425 doi: 10.1111/j.1469-0691.2006.01377.x
See also
Examples
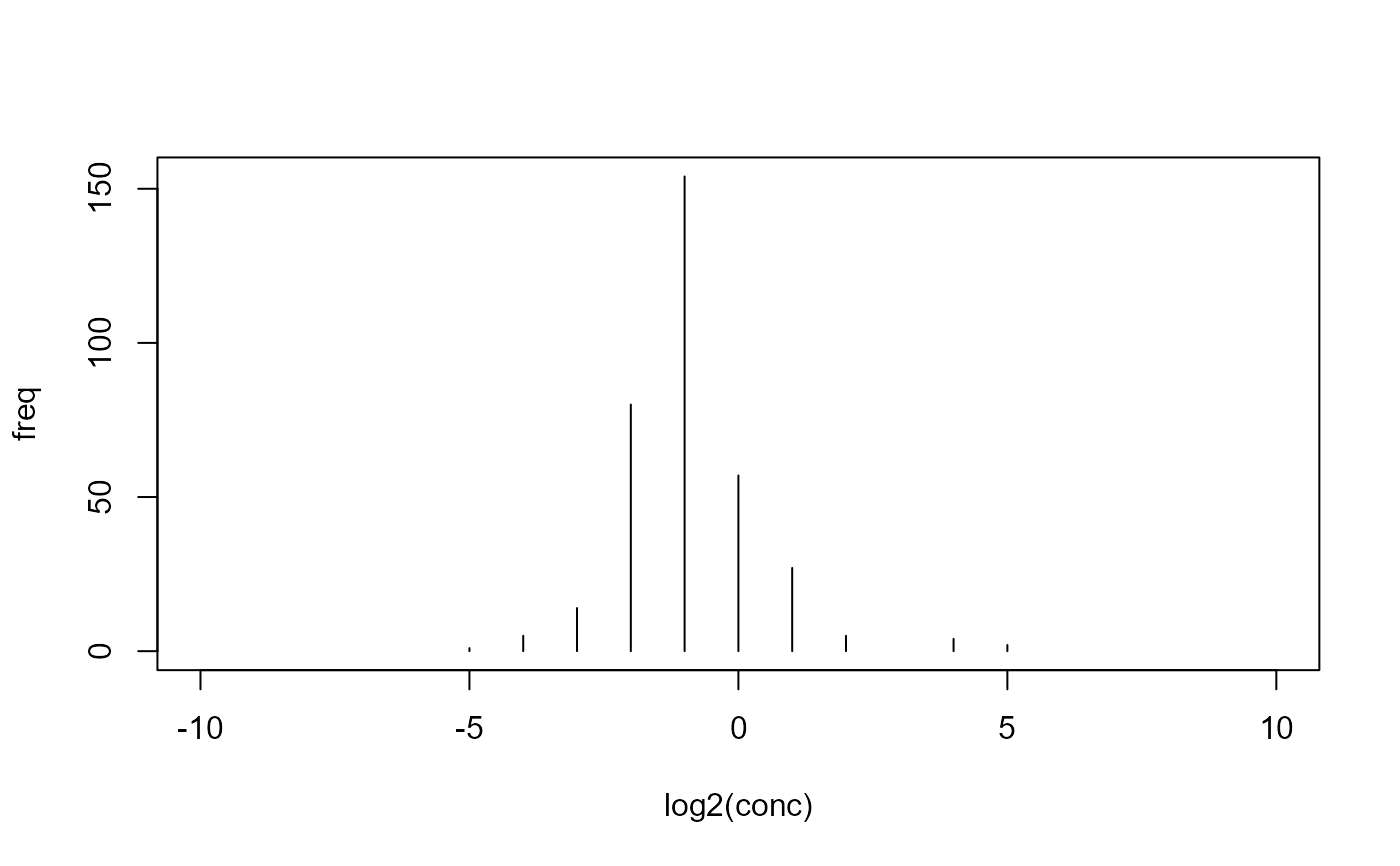
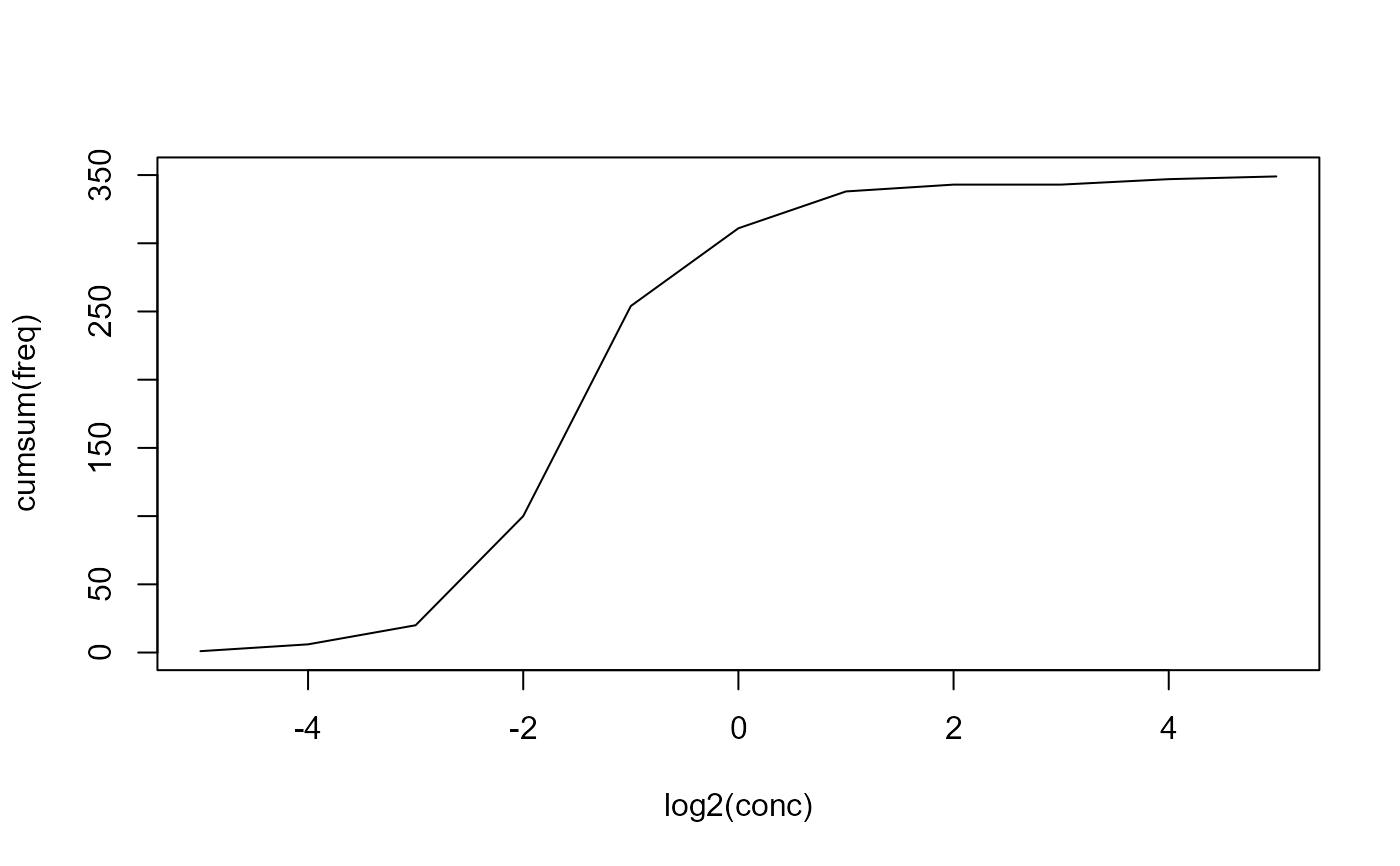
## discard NA values measured <- na.omit(micdata) ## cumulative plot plot(cumsum(freq) ~ log2(conc), data=measured, type="l")x <- log2(measured$conc) y <- measured$freq ## heuristic start values (pstart <- ecoffinder_startpar(x, y, method = "mean"))#> mean sd K #> -0.9111748 1.5165675 349.0000000(pstart <- ecoffinder_startpar(x, y, method = "mode"))#> mean sd K #> -2.010891 1.438260 349.000000(pstart <- ecoffinder_startpar(x, y, method = "peak1"))#> mean sd K #> -1.002944 1.150871 338.000000